Transgenic mice with an R342X mutation in Phf6 display clinical features of Börjeson-Forssman-Lehmann Syndrome
- PMID: 33772537
- PMCID: PMC8120135
- DOI: 10.1093/hmg/ddab081
Transgenic mice with an R342X mutation in Phf6 display clinical features of Börjeson-Forssman-Lehmann Syndrome
Abstract
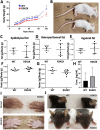
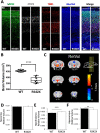
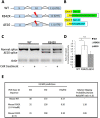
The PHF6 mutation c.1024C > T; p.R342X, is a recurrent cause of Börjeson-Forssman-Lehmann Syndrome (BFLS), a neurodevelopmental disorder characterized by moderate-severe intellectual disability, truncal obesity, gynecomastia, hypogonadism, long tapering fingers and large ears (MIM#301900). Here, we generated transgenic mice with the identical substitution (R342X mice) using CRISPR technology. We show that the p.R342X mutation causes a reduction in PHF6 protein levels, in both human and mice, from nonsense-mediated decay and nonsense-associated alternative splicing, respectively. Magnetic resonance imaging studies indicated that R342X mice had a reduced brain volume on a mixed genetic background but developed hydrocephaly and a high incidence of postnatal death on a C57BL/6 background. Cortical development proceeded normally, while hippocampus and hypothalamus relative brain volumes were altered. A hypoplastic anterior pituitary was also observed that likely contributes to the small size of the R342X mice. Behavior testing demonstrated deficits in associative learning, spatial memory and an anxiolytic phenotype. Taken together, the R342X mice represent a good preclinical model of BFLS that will allow further dissection of PHF6 function and disease pathogenesis.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures





References
-
- Borjeson, M., Forssman, H. and Lehmann, O. (1962) An X-linked, recessively inherited syndrome characterized by grave mental deficiency, epilepsy, and endocrine disorder. Acta Med. Scand., 171, 13–21. - PubMed
-
- Turner, G., Gedeon, A., Mulley, J., Sutherland, G., Rae, J., Power, K. and Arthur, I. (1989) Borjeson-Forssman-Lehmann syndrome: clinical manifestations and gene localization to Xq26-27. Am. J. Med. Genet., 34, 463–469. - PubMed
-
- Visootsak, J., Rosner, B., Dykens, E., Schwartz, C., Hahn, K., White, S.M., Szeftel, R. and Graham, J.M. (2004) Clinical and behavioral features of patients with Borjeson-Forssman-Lehmann syndrome with mutations in PHF6. J. Pediatr., 145, 819–825. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

