Transcribed germline-limited coding sequences in Oxytricha trifallax
- PMID: 33772542
- PMCID: PMC8495736
- DOI: 10.1093/g3journal/jkab092
Transcribed germline-limited coding sequences in Oxytricha trifallax
Abstract
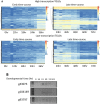
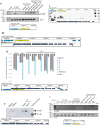
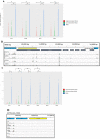
The germline-soma divide is a fundamental distinction in developmental biology, and different genes are expressed in germline and somatic cells throughout metazoan life cycles. Ciliates, a group of microbial eukaryotes, exhibit germline-somatic nuclear dimorphism within a single cell with two different genomes. The ciliate Oxytricha trifallax undergoes massive RNA-guided DNA elimination and genome rearrangement to produce a new somatic macronucleus (MAC) from a copy of the germline micronucleus (MIC). This process eliminates noncoding DNA sequences that interrupt genes and also deletes hundreds of germline-limited open reading frames (ORFs) that are transcribed during genome rearrangement. Here, we update the set of transcribed germline-limited ORFs (TGLOs) in O. trifallax. We show that TGLOs tend to be expressed during nuclear development and then are absent from the somatic MAC. We also demonstrate that exposure to synthetic RNA can reprogram TGLO retention in the somatic MAC and that TGLO retention leads to transcription outside the normal developmental program. These data suggest that TGLOs represent a group of developmentally regulated protein-coding sequences whose gene expression is terminated by DNA elimination.
Keywords: DNA elimination; ciliate; genome rearrangement; germline; micronucleus; noncoding RNA.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
