Immune modulation underpins the anti-cancer activity of HDAC inhibitors
- PMID: 33773029
- PMCID: PMC8637571
- DOI: 10.1002/1878-0261.12953
Immune modulation underpins the anti-cancer activity of HDAC inhibitors
Abstract
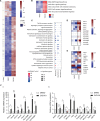
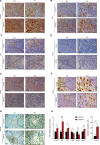
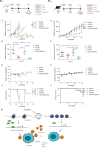
Aberrant protein acetylation is strongly linked to tumorigenesis, and modulating acetylation through targeting histone deacetylase (HDAC) with small-molecule inhibitors has been the focus of clinical trials. However, clinical success on solid tumours, such as colorectal cancer (CRC), has been limited, in part because the cancer-relevant mechanisms through which HDAC inhibitors act remain largely unknown. Here, we have explored, at the genome-wide expression level, the effects of a novel HDAC inhibitor CXD101. In human CRC cell lines, a diverse set of differentially expressed genes were up- and downregulated upon CXD101 treatment. Functional profiling of the expression data highlighted immune-relevant concepts related to antigen processing and natural killer cell-mediated cytotoxicity. Similar profiles were apparent when gene expression was investigated in murine colon26 CRC cells treated with CXD101. Significantly, these changes were also apparent in syngeneic colon26 tumours growing in vivo. The ability of CXD101 to affect immune-relevant gene expression coincided with changes in the tumour microenvironment (TME), especially in the subgroups of CD4 and CD8 tumour-infiltrating T lymphocytes. The altered TME reflected enhanced antitumour activity when CXD101 was combined with immune checkpoint inhibitors (ICIs), such as anti-PD-1 and anti-CTLA4. The ability of CXD101 to reinstate immune-relevant gene expression in the TME and act together with ICIs provides a powerful rationale for exploring the combination therapy in human cancers.
Keywords: HDAC inhibitors; checkpoints inhibitors; immunotherapy; tumour microenvironment.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

