Molecular characterization of interactions between the D614G variant of SARS-CoV-2 S-protein and neutralizing antibodies: A computational approach
- PMID: 33774178
- PMCID: PMC7987576
- DOI: 10.1016/j.meegid.2021.104815
Molecular characterization of interactions between the D614G variant of SARS-CoV-2 S-protein and neutralizing antibodies: A computational approach
Abstract
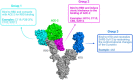
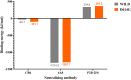
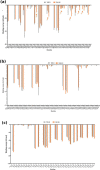
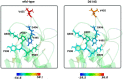
The D614G variant of SARS-CoV-2 S-protein emerged in early 2020 and quickly became the dominant circulating strain in Europe and its environs. The variant was characterized by the higher viral load, which is not associated with disease severity, higher incorporation into the virion, and high cell entry via ACE-2 and TMPRSS2. Previous strains of the coronavirus and the current SARS-CoV-2 have demonstrated the selection of mutations as a mechanism of escaping immune responses. In this study, we used molecular dynamics simulation and MM-PBSA binding energy analysis to provide insights into the behaviour of the D614G S-protein at the molecular level and describe the neutralization mechanism of this variant. Our results show that the D614G S-protein adopts distinct conformational dynamics which is skewed towards the open-state conformation more than the closed-state conformation of the wild-type S-protein. Residue-specific variation of amino acid flexibility and domain-specific RMSD suggest that the mutation causes an allosteric conformational change in the RBD. Evaluation of the interaction energies between the S-protein and neutralizing antibodies show that the mutation may enhance, reduce or not affect the neutralizing interactions depending on the neutralizing antibody, especially if it targets the RBD. The results of this study have shed insights into the behaviour of the D614G S-protein at the molecular level and provided a glimpse of the neutralization mechanism of this variant.
Keywords: D614G; Molecular dynamics simulation; Neutralizing antibody; S-protein; SARS-CoV-2.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare they have no competing interest.
Figures




Similar articles
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization.J Biol Chem. 2021 Jan-Jun;296:100536. doi: 10.1016/j.jbc.2021.100536. Epub 2021 Mar 11. J Biol Chem. 2021. PMID: 33716040 Free PMC article.
-
Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies.Structure. 2021 Jul 1;29(7):655-663.e4. doi: 10.1016/j.str.2021.05.014. Epub 2021 Jun 9. Structure. 2021. PMID: 34111408 Free PMC article.
-
Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection.J Biol Chem. 2021 Jan-Jun;296:100135. doi: 10.1074/jbc.REV120.015980. Epub 2020 Dec 6. J Biol Chem. 2021. PMID: 33268377 Free PMC article. Review.
-
Why are some coronavirus variants more infectious?J Biosci. 2021;46(4):101. doi: 10.1007/s12038-021-00221-y. J Biosci. 2021. PMID: 34785628 Free PMC article. Review.
Cited by
-
SARS-CoV-2 spike glycoprotein-reactive T cells can be readily expanded from COVID-19 vaccinated donors.Immun Inflamm Dis. 2021 Dec;9(4):1452-1467. doi: 10.1002/iid3.496. Epub 2021 Jul 27. Immun Inflamm Dis. 2021. PMID: 34314576 Free PMC article.
-
N-terminal domain of SARS CoV-2 spike protein mutation associated reduction in effectivity of neutralizing antibody with vaccinated individuals.J Med Virol. 2021 Oct;93(10):5726-5728. doi: 10.1002/jmv.27181. Epub 2021 Jul 14. J Med Virol. 2021. PMID: 34232521 Free PMC article. No abstract available.
-
Novel coronavirus mutations: Vaccine development and challenges.Microb Pathog. 2022 Dec;173(Pt A):105828. doi: 10.1016/j.micpath.2022.105828. Epub 2022 Oct 13. Microb Pathog. 2022. PMID: 36243381 Free PMC article. Review.
-
Impact of mutations in SARS-COV-2 spike on viral infectivity and antigenicity.Brief Bioinform. 2022 Jan 17;23(1):bbab375. doi: 10.1093/bib/bbab375. Brief Bioinform. 2022. PMID: 34518867 Free PMC article.
-
The influence of single-point mutation D614G on the binding process between human angiotensin-converting enzyme 2 and the SARS-CoV-2 spike protein-an atomistic simulation study.RSC Adv. 2023 Mar 28;13(15):9800-9810. doi: 10.1039/d3ra00198a. eCollection 2023 Mar 27. RSC Adv. 2023. PMID: 36998522 Free PMC article.
References
-
- Abraham M.J., et al. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25.
-
- Barnes C.O., West A.P.J., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., Lorenzi J.C.C., Finkin S., Hägglöf T., Hurley A., Millard K.G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P.D.…Bjorkman P.J. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182(4):828–842. doi: 10.1016/j.cell.2020.06.025. e16. - DOI - PMC - PubMed
-
- Beltrán-Pavez C., Riquelme-Barrios S., Oyarzún-Arrau A., Gaete-Argel A., González-Stegmaier R., Cereceda-Solis K., Aguirre A., Travisany D., Palma-Vejares R., Barriga G.P., Gaggero A., Martínez-Valdebenito C., Le Corre N., Ferrés M., Balcells M.E., Fernandez J., Ramírez E., Villarroel F., Valiente-Echeverría F., Soto-Rifo R. Insights into neutralizing antibody responses in individuals exposed to SARS-CoV-2 in Chile. Sci. Adv. 2021;7(7) doi: 10.1126/sciadv.abe6855. - DOI - PMC - PubMed
-
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L.…Chen W. Vol. 655. 2020. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2; pp. 650–655. August. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

