Taxogenomic and Comparative Genomic Analysis of the Genus Saccharomonospora Focused on the Identification of Biosynthetic Clusters PKS and NRPS
- PMID: 33776952
- PMCID: PMC7990883
- DOI: 10.3389/fmicb.2021.603791
Taxogenomic and Comparative Genomic Analysis of the Genus Saccharomonospora Focused on the Identification of Biosynthetic Clusters PKS and NRPS
Abstract
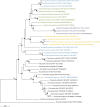
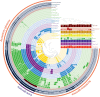
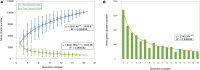
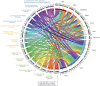
Actinobacteria are prokaryotes with a large biotechnological interest due to their ability to produce secondary metabolites, produced by two main biosynthetic gene clusters (BGCs): polyketide synthase (PKS) and non-ribosomal peptide synthetase (NRPS). Most studies on bioactive products have been carried out on actinobacteria isolated from soil, freshwater or marine habitats, while very few have been focused on halophilic actinobacteria isolated from extreme environments. In this study we have carried out a comparative genomic analysis of the actinobacterial genus Saccharomonospora, which includes species isolated from soils, lake sediments, marine or hypersaline habitats. A total of 19 genome sequences of members of Saccharomonospora were retrieved and analyzed. We compared the 16S rRNA gene-based phylogeny of this genus with evolutionary relationships inferred using a phylogenomic approach obtaining almost identical topologies between both strategies. This method allowed us to unequivocally assign strains into species and to identify some taxonomic relationships that need to be revised. Our study supports a recent speciation event occurring between Saccharomonospora halophila and Saccharomonospora iraqiensis. Concerning the identification of BGCs, a total of 18 different types of BGCs were detected in the analyzed genomes of Saccharomonospora, including PKS, NRPS and hybrid clusters which might be able to synthetize 40 different putative products. In comparison to other genera of the Actinobacteria, members of the genus Saccharomonospora showed a high degree of novelty and diversity of BGCs.
Keywords: Saccharomonospora; actinobacteria; biosynthetic gene clusters; comparative genomic analysis; non-ribosomal peptide synthetase; polyketide synthase; secondary metabolites; taxophylogenomic analysis.
Copyright © 2021 Ramírez-Durán, de la Haba, Vera-Gargallo, Sánchez-Porro, Alonso-Carmona, Sandoval-Trujillo and Ventosa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Borriss R., Rueckert C., Blom J., Bezuidt O., Reva O., Klenk H.-P. (2011). “Whole genome sequence comparisons in taxonomy,” in Methods in Microbiology. Taxonomy of Prokaryotes, eds Rainey F., Oren A. (London: Academic Press; ), 409–436. 10.1016/B978-0-12-387730-7.00018-8 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

