T Cell Activation, Highly Armed Cytotoxic Cells and a Shift in Monocytes CD300 Receptors Expression Is Characteristic of Patients With Severe COVID-19
- PMID: 33777054
- PMCID: PMC7991729
- DOI: 10.3389/fimmu.2021.655934
T Cell Activation, Highly Armed Cytotoxic Cells and a Shift in Monocytes CD300 Receptors Expression Is Characteristic of Patients With Severe COVID-19
Abstract
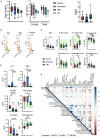
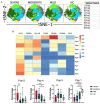
COVID-19 manifests with a wide diversity of clinical phenotypes characterized by dysfunctional and exaggerated host immune responses. Many results have been described on the status of the immune system of patients infected with SARS-CoV-2, but there are still aspects that have not been fully characterized or understood. In this study, we have analyzed a cohort of patients with mild, moderate and severe disease. We performed flow cytometric studies and correlated the data with the clinical characteristics and clinical laboratory values of the patients. Both conventional and unsupervised data analyses concluded that patients with severe disease are characterized, among others, by a higher state of activation in all T cell subsets (CD4, CD8, double negative and T follicular helper cells), higher expression of perforin and granzyme B in cytotoxic cells, expansion of adaptive NK cells and the accumulation of activated and immature dysfunctional monocytes which are identified by a low expression of HLA-DR and an intriguing shift in the expression pattern of CD300 receptors. More importantly, correlation analysis showed a strong association between the alterations in the immune cells and the clinical signs of severity. These results indicate that patients with severe COVID-19 have a broad perturbation of their immune system, and they will help to understand the immunopathogenesis of COVID-19.
Keywords: CD300a; CD300c; CD300e; COVID-19; NK cells; T cells; granzyme B; monocytes.
Copyright © 2021 Zenarruzabeitia, Astarloa-Pando, Terrén, Orrantia, Pérez-Garay, Seijas-Betolaza, Nieto-Arana, Imaz-Ayo, Pérez-Fernández, Arana-Arri and Borrego.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

