The oralome and its dysbiosis: New insights into oral microbiome-host interactions
- PMID: 33777334
- PMCID: PMC7960681
- DOI: 10.1016/j.csbj.2021.02.010
The oralome and its dysbiosis: New insights into oral microbiome-host interactions
Abstract
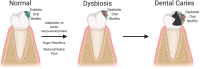
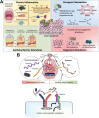
The oralome is the summary of the dynamic interactions orchestrated between the ecological community of oral microorganisms (comprised of up to approximately 1000 species of bacteria, fungi, viruses, archaea and protozoa - the oral microbiome) that live in the oral cavity and the host. These microorganisms form a complex ecosystem that thrive in the dynamic oral environment in a symbiotic relationship with the human host. However, the microbial composition is significantly affected by interspecies and host-microbial interactions, which in turn, can impact the health and disease status of the host. In this review, we discuss the composition of the oralome and inter-species and host-microbial interactions that take place in the oral cavity and examine how these interactions change from healthy (eubiotic) to disease (dysbiotic) states. We further discuss the dysbiotic signatures associated with periodontitis and caries and their sequalae, (e.g., tooth/bone loss and pulpitis), and the systemic diseases associated with these oral diseases, such as infective endocarditis, atherosclerosis, diabetes, Alzheimer's disease and head and neck/oral cancer. We then discuss current computational techniques to assess dysbiotic oral microbiome changes. Lastly, we discuss current and novel techniques for modulation of the dysbiotic oral microbiome that may help in disease prevention and treatment, including standard hygiene methods, prebiotics, probiotics, use of nano-sized drug delivery systems (nano-DDS), extracellular polymeric matrix (EPM) disruption, and host response modulators.
Keywords: Complex diseases; Dysbiosis; Host-microbe interactions; Oral biofilm; Oral microbiome.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Berg R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. - PubMed
-
- Dovrolis N., Kolios G., Spyrou G.M., Maroulakou I. Computational profiling of the gut-brain axis: microflora dysbiosis insights to neurological disorders. Brief Bioinf. 2019;20:825–841. - PubMed
-
- Proal A.D., Lindseth I.A., Marshall T.G. Microbe-microbe and host-microbe interactions drive microbiome dysbiosis and inflammatory processes. Discov Med. 2017;23:51–60. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
