Diversity of the reaction mechanisms of SAM-dependent enzymes
- PMID: 33777672
- PMCID: PMC7982431
- DOI: 10.1016/j.apsb.2020.08.011
Diversity of the reaction mechanisms of SAM-dependent enzymes
Abstract
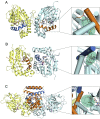
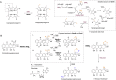
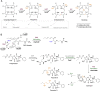
S-adenosylmethionine (SAM) is ubiquitous in living organisms and is of great significance in metabolism as a cofactor of various enzymes. Methyltransferases (MTases), a major group of SAM-dependent enzymes, catalyze methyl transfer from SAM to C, O, N, and S atoms in small-molecule secondary metabolites and macromolecules, including proteins and nucleic acids. MTases have long been a hot topic in biomedical research because of their crucial role in epigenetic regulation of macromolecules and biosynthesis of natural products with prolific pharmacological moieties. However, another group of SAM-dependent enzymes, sharing similar core domains with MTases, can catalyze nonmethylation reactions and have multiple functions. Herein, we mainly describe the nonmethylation reactions of SAM-dependent enzymes in biosynthesis. First, we compare the structural and mechanistic similarities and distinctions between SAM-dependent MTases and the non-methylating SAM-dependent enzymes. Second, we summarize the reactions catalyzed by these enzymes and explore the mechanisms. Finally, we discuss the structural conservation and catalytical diversity of class I-like non-methylating SAM-dependent enzymes and propose a possibility in enzymes evolution, suggesting future perspectives for enzyme-mediated chemistry and biotechnology, which will help the development of new methods for drug synthesis.
Keywords: Biocatalysis; Catalytic mechanism; Methyltransferase; Nonmethylation reaction; SAM-dependent enzyme.
© 2021 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures

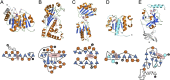
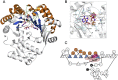


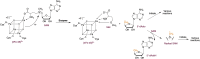
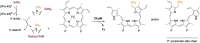




References
-
- Cantoni G.L. Biological methylation: selected aspects. Annu Rev Biochem. 1975;44:435–451. - PubMed
-
- Chen H., Wang Z., Cai H., Zhou C. Progress in the microbial production of S-adenosyl-l-methionine. World J Microbiol Biotechnol. 2016;32:153. - PubMed
-
- He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. - PubMed
-
- Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

