The Effects of Autophagy-Related Genes and lncRNAs in Therapy and Prognosis of Colorectal Cancer
- PMID: 33777735
- PMCID: PMC7991845
- DOI: 10.3389/fonc.2021.582040
The Effects of Autophagy-Related Genes and lncRNAs in Therapy and Prognosis of Colorectal Cancer
Abstract
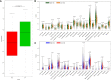
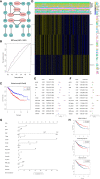
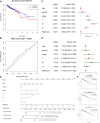
Cellular autophagy plays an important role in the occurrence and development of colorectal cancer (CRC). Whether autophagy-related genes and lncRNAs can be used as ideal markers in CRC is still controversial. The purpose of this study is to identify novel treatment and prognosis markers of CRC. We downloaded transcription and clinical data of CRC from the GEO (GSE40967, GSE12954, GSE17536) and TCGA database, screened for differentially autophagy-related genes (DEAGs) and lncRNAs, constructed prognostic model, and analyzed its relationship with immune infiltration. TCGA and GEO datasets (GSE12954 and GSE17536) were used to validate the effect of the model. Oncomine database and Human Protein Atlas verified the expression of DEAGs. We obtained a total of 151 DEAGs in three verification sets collaboratively. Then we constructed a risk prognostic model through Lasso regression to obtain 15 prognostic DEAGs from the training set and verified the risk prognostic model in three verification sets. The low-risk group survived longer than the high-risk group. Age, gender, pathological stage, and TNM stage were related to the prognostic risk of CRC. On the other hand, BRAF status, RFS event, and tumor location are considered as most significant risk factors of CRC in the training set. Furthermore, we found that the immune score of the low-risk group was higher. The content of CD8 + T cells, active NK cells, macrophages M0, macrophages M1, and active dendritic cells was noted more in the high-risk group. The content of plasma cells, resting memory CD4 + T cells, resting NK cells, resting mast cells, and neutrophil cells was higher in the low-risk group. After all, the Oncomine database and immunohistochemistry verified that the expression level of most key autophagy-related genes was consistent with the results that we found. In addition, we obtained six lncRNAs co-expressed with DEAGs from the training set and found that the survival time was longer in the low-risk group. This finding was verified in the verification set and showed same trend to the results mentioned above. In the final analysis, these results indicate that autophagy-related genes and lncRNAs can be used as prognostic and therapeutic markers for CRC.
Keywords: autophagy; colorectal cancer; gene; immune; lncRNA; prognosis.
Copyright © 2021 Yang, Feng, Bai, Zhang, Zhou, Liao, Lei, Zhang, Huang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

