Aberrations of Genomic Imprinting in Glioblastoma Formation
- PMID: 33777782
- PMCID: PMC7994891
- DOI: 10.3389/fonc.2021.630482
Aberrations of Genomic Imprinting in Glioblastoma Formation
Abstract
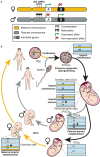
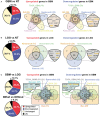
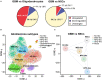
In human glioblastoma (GBM), the presence of a small population of cells with stem cell characteristics, the glioma stem cells (GSCs), has been described. These cells have GBM potential and are responsible for the origin of the tumors. However, whether GSCs originate from normal neural stem cells (NSCs) as a consequence of genetic and epigenetic changes and/or dedifferentiation from somatic cells remains to be investigated. Genomic imprinting is an epigenetic marking process that causes genes to be expressed depending on their parental origin. The dysregulation of the imprinting pattern or the loss of genomic imprinting (LOI) have been described in different tumors including GBM, being one of the earliest and most common events that occurs in human cancers. Here we have gathered the current knowledge of the role of imprinted genes in normal NSCs function and how the imprinting process is altered in human GBM. We also review the changes at particular imprinted loci that might be involved in the development of the tumor. Understanding the mechanistic similarities in the regulation of genomic imprinting between normal NSCs and GBM cells will be helpful to identify molecular players that might be involved in the development of human GBM.
Keywords: genomic imprinting; glioblastoma; methylation; neural stem cells; subventricular zone.
Copyright © 2021 Lozano-Ureña, Jiménez-Villalba, Pinedo-Serrano, Jordán-Pla, Kirstein and Ferrón.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

