Spatial mapping of SARS-CoV-2 and H1N1 lung injury identifies differential transcriptional signatures
- PMID: 33778787
- PMCID: PMC7985929
- DOI: 10.1016/j.xcrm.2021.100242
Spatial mapping of SARS-CoV-2 and H1N1 lung injury identifies differential transcriptional signatures
Abstract
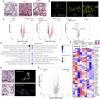
Severe SARS-CoV-2 infection often leads to the development of acute respiratory distress syndrome (ARDS), with profound pulmonary patho-histological changes post-mortem. It is not clear whether ARDS from SARS-CoV-2 is similar to that observed in influenza H1N1, another common viral cause of lung injury. Here, we analyze specific ARDS regions of interest utilizing a spatial transcriptomic platform on autopsy-derived lung tissue from patients with SARS-CoV-2 (n = 3), H1N1 (n = 3), and a dual infected individual (n = 1). Enhanced gene signatures in alveolar epithelium, vascular tissue, and lung macrophages identify not only increased regional coagulopathy but also increased extracellular remodeling, alternative macrophage activation, and squamous metaplasia of type II pneumocytes in SARS-CoV-2. Both the H1N1 and dual-infected transcriptome demonstrated an enhanced antiviral response compared to SARS-CoV-2. Our results uncover regional transcriptional changes related to tissue damage/remodeling, altered cellular phenotype, and vascular injury active in SARS-CoV-2 and present therapeutic targets for COVID-19-related ARDS.
Keywords: ARDS; COVID-19; H1N1 influenza; SARS-CoV-2; spatial transcriptomics.
© 2021 The Author(s).
Conflict of interest statement
K.T., Y.L., and L.Z. are employees of Nanostring technologies. There is no financial conflict of interest to disclose for this study (all services provided by Nanostring technologies were paid under the grants awarded to the senior author).
Figures




References
-
- Auld S.C., Caridi-Scheible M., Blum J.M., Robichaux C., Kraft C., Jacob J.T., Jabaley C.S., Carpenter D., Kaplow R., Hernandez-Romieu A.C., Emory COVID-19 Quality and Clinical Research Collaborative ICU and Ventilator Mortality Among Critically Ill Adults With Coronavirus Disease 2019. Crit. Care Med. 2020;48:e799–e804. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

