Ubiquitin-dependent regulation of transcription in development and disease
- PMID: 33779035
- PMCID: PMC8025022
- DOI: 10.15252/embr.202051078
Ubiquitin-dependent regulation of transcription in development and disease
Abstract
Transcription is an elaborate process that is required to establish and maintain the identity of the more than two hundred cell types of a metazoan organism. Strict regulation of gene expression is therefore vital for tissue formation and homeostasis. An accumulating body of work found that ubiquitylation of histones, transcription factors, or RNA polymerase II is crucial for ensuring that transcription occurs at the right time and place during development. Here, we will review principles of ubiquitin-dependent control of gene expression and discuss how breakdown of these regulatory circuits leads to a wide array of human diseases.
Keywords: RNA polymerase II; histone modification; transcription; ubiquitin.
© 2021 The Authors.
Conflict of interest statement
M.R. is a co‐founder and consultant to Nurix and scientific advisory board member to Monte Rosa.
Figures

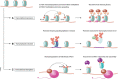

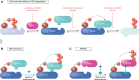
References
-
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S et al (2005) The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123: 409–421 - PubMed
-
- Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, Mayer S, Takahashi S, Licht JD, Prive GG (2003) Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell 12: 1551–1564 - PubMed
-
- Anindya R, Aygun O, Svejstrup JQ (2007) Damage‐induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell 28: 386–397 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

