Translational activity is uncoupled from nucleic acid content in bacterial cells of the human gut microbiota
- PMID: 33779505
- PMCID: PMC8009119
- DOI: 10.1080/19490976.2021.1903289
Translational activity is uncoupled from nucleic acid content in bacterial cells of the human gut microbiota
Abstract
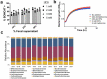
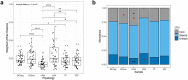
Changes in bacterial diversity in the human gut have been associated with many conditions, despite not always reflecting changes in bacterial activity. Methods linking bacterial identity to function are needed for improved understanding of how bacterial communities adapt and respond to their environment, including the gut. Here, we optimized bioorthogonal non-canonical amino acid tagging (BONCAT) for the gut microbiota and combined it with fluorescently activated cell sorting and sequencing (FACS-Seq) to identify the translationally active members of the community. We then used this novel technique to compare with other bulk community measurements of activity and viability: relative nucleic acid content and membrane damage. The translationally active bacteria represent about half of the gut microbiota, and are not distinct from the whole community. The high nucleic acid content bacteria also represent half of the gut microbiota, but are distinct from the whole community and correlate with the damaged subset. Perturbing the community with xenobiotics previously shown to alter bacterial activity but not diversity resulted in stronger changes in the distinct physiological fractions than in the whole community. BONCAT is a suitable method to probe the translationally active members of the gut microbiota, and combined with FACS-Seq, allows for their identification. The high nucleic acid content bacteria are not necessarily the protein-producing bacteria in the community; thus, further work is needed to understand the relationship between nucleic acid content and bacterial metabolism in the human gut. Considering physiologically distinct subsets of the gut microbiota may be more informative than whole-community profiling.
Keywords: BONCAT; Gut microbiome; bacterial activity; bacterial physiology; flow cytometry; single-cell.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nat. 2016;535:94–103. [Internet]. - PubMed
-
- Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli M, et al. Differences in fecal microbiota in different european study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72(2):1027. doi: 10.1128/AEM.72.2.1027-1033.2006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
