An atlas of the binding specificities of transcription factors in Pseudomonas aeruginosa directs prediction of novel regulators in virulence
- PMID: 33779544
- PMCID: PMC8041468
- DOI: 10.7554/eLife.61885
An atlas of the binding specificities of transcription factors in Pseudomonas aeruginosa directs prediction of novel regulators in virulence
Abstract
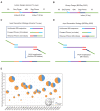
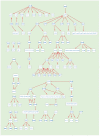
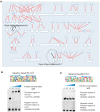
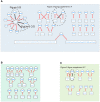
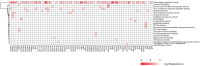
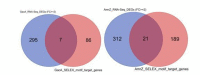
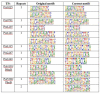
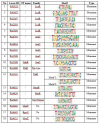
A high-throughput systematic evolution of ligands by exponential enrichment assay was applied to 371 putative TFs in Pseudomonas aeruginosa, which resulted in the robust enrichment of 199 unique sequence motifs describing the binding specificities of 182 TFs. By scanning the genome, we predicted in total 33,709 significant interactions between TFs and their target loci, which were more than 11-fold enriched in the intergenic regions but depleted in the gene body regions. To further explore and delineate the physiological and pathogenic roles of TFs in P. aeruginosa, we constructed regulatory networks for nine major virulence-associated pathways and found that 51 TFs were potentially significantly associated with these virulence pathways, 32 of which had not been characterized before, and some were even involved in multiple pathways. These results will significantly facilitate future studies on transcriptional regulation in P. aeruginosa and other relevant pathogens, and accelerate to discover effective treatment and prevention strategies for the associated infectious diseases.
Keywords: Pseudomonas aeruginosa; dna binding specificity; gene expression; genetics; genomics; infectious disease; microbiology; transcription factors; transcriptional regulation network; virulence.
© 2021, Wang et al.
Conflict of interest statement
TW, WS, LF, CH, NW, SF, JZ, XD, JY No competing interests declared
Figures


















References
-
- Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Research. 2012;40:7207–7218. doi: 10.1093/nar/gks384. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- 8187364/National Natural Science Foundation of China
- 7005314/City University of Hong Kong
- 31900443/National Natural Science Foundation of China
- 31870116/National Natural Science Foundation of China
- 21103018/Research Grants Council, University Grants Committee
- 21100420/Research Grants Council, University Grants Committee
- 11101619/Research Grants Council, University Grants Committee
- 2019M663799/China Postdoctoral Science Foundation
- 2019M663794/China Postdoctoral Science Foundation
- 9667188/City University of Hong Kong
- 32070596/National Natural Science Foundation of China
- 9610424/City University of Hong Kong
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

