Inhibition of macrophage histone demethylase JMJD3 protects against abdominal aortic aneurysms
- PMID: 33779682
- PMCID: PMC8008365
- DOI: 10.1084/jem.20201839
Inhibition of macrophage histone demethylase JMJD3 protects against abdominal aortic aneurysms
Abstract
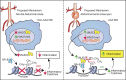
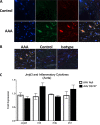
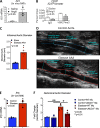
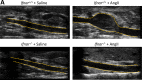
Abdominal aortic aneurysms (AAAs) are a life-threatening disease for which there is a lack of effective therapy preventing aortic rupture. During AAA formation, pathological vascular remodeling is driven by macrophage infiltration, and the mechanisms regulating macrophage-mediated inflammation remain undefined. Recent evidence suggests that an epigenetic enzyme, JMJD3, plays a critical role in establishing macrophage phenotype. Using single-cell RNA sequencing of human AAA tissues, we identified increased JMJD3 in aortic monocyte/macrophages resulting in up-regulation of an inflammatory immune response. Mechanistically, we report that interferon-β regulates Jmjd3 expression via JAK/STAT and that JMJD3 induces NF-κB-mediated inflammatory gene transcription in infiltrating aortic macrophages. In vivo targeted inhibition of JMJD3 with myeloid-specific genetic depletion (JMJD3f/fLyz2Cre+) or pharmacological inhibition in the elastase or angiotensin II-induced AAA model preserved the repressive H3K27me3 on inflammatory gene promoters and markedly reduced AAA expansion and attenuated macrophage-mediated inflammation. Together, our findings suggest that cell-specific pharmacologic therapy targeting JMJD3 may be an effective intervention for AAA expansion.
© 2021 Davis et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures






Comment in
-
Immune cells-A curse and a blessing!J Exp Med. 2021 Jun 7;218(6):e20210590. doi: 10.1084/jem.20210590. Epub 2021 May 21. J Exp Med. 2021. PMID: 34019638 Free PMC article.
Similar articles
-
Palmitate-TLR4 signaling regulates the histone demethylase, JMJD3, in macrophages and impairs diabetic wound healing.Eur J Immunol. 2020 Dec;50(12):1929-1940. doi: 10.1002/eji.202048651. Epub 2020 Jul 20. Eur J Immunol. 2020. PMID: 32662520 Free PMC article.
-
The Histone Methyltransferase SETDB2 Modulates Tissue Inhibitors of Metalloproteinase-Matrix Metalloproteinase Activity During Abdominal Aortic Aneurysm Development.Ann Surg. 2023 Sep 1;278(3):426-440. doi: 10.1097/SLA.0000000000005963. Epub 2023 Jun 16. Ann Surg. 2023. PMID: 37325923 Free PMC article.
-
Epigenetic Modulation in Periodontitis: Interaction of Adiponectin and JMJD3-IRF4 Axis in Macrophages.J Cell Physiol. 2016 May;231(5):1090-6. doi: 10.1002/jcp.25201. Epub 2015 Oct 6. J Cell Physiol. 2016. PMID: 26399931 Free PMC article.
-
Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process.J Mol Med (Berl). 2014 Oct;92(10):1035-43. doi: 10.1007/s00109-014-1182-x. Epub 2014 Jun 14. J Mol Med (Berl). 2014. PMID: 24925089 Review.
-
The Role of Epigenetic Modifications in Abdominal Aortic Aneurysm Pathogenesis.Biomolecules. 2022 Jan 21;12(2):172. doi: 10.3390/biom12020172. Biomolecules. 2022. PMID: 35204673 Free PMC article. Review.
Cited by
-
NINJ1 Facilitates Abdominal Aortic Aneurysm Formation via Blocking TLR4-ANXA2 Interaction and Enhancing Macrophage Infiltration.Adv Sci (Weinh). 2024 Aug;11(31):e2306237. doi: 10.1002/advs.202306237. Epub 2024 Jun 23. Adv Sci (Weinh). 2024. PMID: 38922800 Free PMC article.
-
Emodin inhibits bladder inflammation and fibrosis in mice with interstitial cystitis by regulating JMJD3.Acta Cir Bras. 2023 Dec 1;38:e385123. doi: 10.1590/acb385123. eCollection 2023. Acta Cir Bras. 2023. PMID: 38055393 Free PMC article.
-
Identification of PTPN22 as a potential genetic biomarker for abdominal aortic aneurysm.Front Cardiovasc Med. 2022 Dec 14;9:1061771. doi: 10.3389/fcvm.2022.1061771. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36588574 Free PMC article.
-
M1 Macrophage-Derived Exosome LncRNA PVT1 Promotes Inflammation and Pyroptosis of Vascular Smooth Muscle Cells in Abdominal Aortic Aneurysm by Inhibiting miR-186-5p and Regulating HMGB1.Cardiovasc Toxicol. 2024 Mar;24(3):302-320. doi: 10.1007/s12012-024-09838-5. Epub 2024 Mar 7. Cardiovasc Toxicol. 2024. PMID: 38453799 Free PMC article.
-
Deficiency of IL-7R attenuates abdominal aortic aneurysms in mice by inhibiting macrophage polarization towards M1 phenotype through the NF-κB pathway.Mol Med. 2025 Apr 16;31(1):138. doi: 10.1186/s10020-025-01209-2. Mol Med. 2025. PMID: 40240976 Free PMC article.
References
-
- Boytard, L., Spear R., Chinetti-Gbaguidi G., Acosta-Martin A.E., Vanhoutte J., Lamblin N., Staels B., Amouyel P., Haulon S., and Pinet F.. 2013. Role of proinflammatory CD68(+) mannose receptor(-) macrophages in peroxiredoxin-1 expression and in abdominal aortic aneurysms in humans. Arterioscler. Thromb. Vasc. Biol. 33:431–438. 10.1161/ATVBAHA.112.300663 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

