Gene Set Knowledge Discovery with Enrichr
- PMID: 33780170
- PMCID: PMC8152575
- DOI: 10.1002/cpz1.90
Gene Set Knowledge Discovery with Enrichr
Abstract
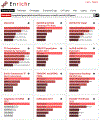
Profiling samples from patients, tissues, and cells with genomics, transcriptomics, epigenomics, proteomics, and metabolomics ultimately produces lists of genes and proteins that need to be further analyzed and integrated in the context of known biology. Enrichr (Chen et al., 2013; Kuleshov et al., 2016) is a gene set search engine that enables the querying of hundreds of thousands of annotated gene sets. Enrichr uniquely integrates knowledge from many high-profile projects to provide synthesized information about mammalian genes and gene sets. The platform provides various methods to compute gene set enrichment, and the results are visualized in several interactive ways. This protocol provides a summary of the key features of Enrichr, which include using Enrichr programmatically and embedding an Enrichr button on any website. © 2021 Wiley Periodicals LLC. Basic Protocol 1: Analyzing lists of differentially expressed genes from transcriptomics, proteomics and phosphoproteomics, GWAS studies, or other experimental studies Basic Protocol 2: Searching Enrichr by a single gene or key search term Basic Protocol 3: Preparing raw or processed RNA-seq data through BioJupies in preparation for Enrichr analysis Basic Protocol 4: Analyzing gene sets for model organisms using modEnrichr Basic Protocol 5: Using Enrichr in Geneshot Basic Protocol 6: Using Enrichr in ARCHS4 Basic Protocol 7: Using the enrichment analysis visualization Appyter to visualize Enrichr results Basic Protocol 8: Using the Enrichr API Basic Protocol 9: Adding an Enrichr button to a website.
Keywords: bioinformatics; disease; drug discovery; enrichment analysis; gene sets; visualization; web application.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT:
The authors declare no conflicts of interest.
Figures



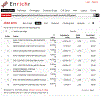











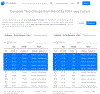







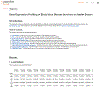




















References
LITERATURE CITED:
KEY REFERENCES:
-
- Clarke DJB, Jeon M, Stein DJ, Moiseyev N, Kropiwnicki E, Dai C, Xie Z, Wojciechowicz ML, Litz S, Hom J, Evangelista JE, Goldman L, Zhang S, Yoon C, Ahamed T, Bhuiyan S, Cheng M, Karam J, Jagodnik JM, Shu I, Lachmann A, Ayling S, Jenkins SL, Ma’ayan A (2021). Appyters: Turning Jupyter Notebooks into Data Driven Web Apps. Patterns (Accepted). - PMC - PubMed
-
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, & Ma’ayan A (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res, 44(W1), W90–97. 10.1093/nar/gkw377 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

