CST in maintaining genome stability: Beyond telomeres
- PMID: 33780718
- PMCID: PMC8081025
- DOI: 10.1016/j.dnarep.2021.103104
CST in maintaining genome stability: Beyond telomeres
Abstract
The human CST (CTC1-STN1-TEN1) complex is an RPA-like single-stranded DNA binding protein complex. While its telomeric functions have been well investigated, numerous studies have revealed that hCST also plays important roles in maintaining genome stability beyond telomeres. Here, we review and discuss recent discoveries on CST in various global genome maintenance pathways, including findings on the CST supercomplex structure, its functions in unperturbed DNA replication, stalled replication, double-strand break repair, and the ATR-CHK1 activation pathway. By summarizing these recent discoveries, we hope to offer new insights into genome maintenance mechanisms and the pathogenesis of CST mutation-associated diseases.
Keywords: CTC1-STN1-TEN1; DNA replication; DSB repair; Fork stalling; Genome stability; Replication stress.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures
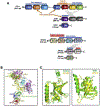
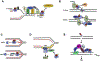
Similar articles
-
Human CTC1 promotes TopBP1 stability and CHK1 phosphorylation in response to telomere dysfunction and global replication stress.Cell Cycle. 2020 Dec;19(24):3491-3507. doi: 10.1080/15384101.2020.1849979. Epub 2020 Dec 3. Cell Cycle. 2020. PMID: 33269665 Free PMC article.
-
Emerging roles of CST in maintaining genome stability and human disease.Front Biosci (Landmark Ed). 2018 Mar 1;23(8):1564-1586. doi: 10.2741/4661. Front Biosci (Landmark Ed). 2018. PMID: 29293451 Free PMC article. Review.
-
Guardians of the Genome: How the Single-Stranded DNA-Binding Proteins RPA and CST Facilitate Telomere Replication.Biomolecules. 2024 Feb 22;14(3):263. doi: 10.3390/biom14030263. Biomolecules. 2024. PMID: 38540683 Free PMC article. Review.
-
Human TEN1 maintains telomere integrity and functions in genome-wide replication restart.J Biol Chem. 2013 Oct 18;288(42):30139-30150. doi: 10.1074/jbc.M113.493478. Epub 2013 Sep 11. J Biol Chem. 2013. PMID: 24025336 Free PMC article.
-
ATR cooperates with CTC1 and STN1 to maintain telomeres and genome integrity in Arabidopsis.Mol Biol Cell. 2012 Apr;23(8):1558-68. doi: 10.1091/mbc.E11-12-1002. Epub 2012 Feb 22. Mol Biol Cell. 2012. PMID: 22357613 Free PMC article.
Cited by
-
The Intrinsically Disordered Region in the Human STN1 OB-Fold Domain Is Important for Protecting Genome Stability.Biology (Basel). 2021 Sep 28;10(10):977. doi: 10.3390/biology10100977. Biology (Basel). 2021. PMID: 34681076 Free PMC article.
-
Structural genomics approach to investigate deleterious impact of nsSNPs in conserved telomere maintenance component 1.Sci Rep. 2021 May 13;11(1):10202. doi: 10.1038/s41598-021-89450-7. Sci Rep. 2021. PMID: 33986331 Free PMC article.
-
Molecular architecture and oligomerization of Candida glabrata Cdc13 underpin its telomeric DNA-binding and unfolding activity.Nucleic Acids Res. 2023 Jan 25;51(2):668-686. doi: 10.1093/nar/gkac1261. Nucleic Acids Res. 2023. PMID: 36629261 Free PMC article.
-
The stn1-sz2 Mutant Provides New Insight into the Impacts of Telomeric Cdc13-Stn1-Ten1 Dysfunction on Cell Cycle Progression.Cells. 2025 May 26;14(11):784. doi: 10.3390/cells14110784. Cells. 2025. PMID: 40497960 Free PMC article.
-
Deficiency in mammalian STN1 promotes colon cancer development via inhibiting DNA repair.Sci Adv. 2023 May 10;9(19):eadd8023. doi: 10.1126/sciadv.add8023. Epub 2023 May 10. Sci Adv. 2023. PMID: 37163605 Free PMC article.
References
-
- Vijg J and Suh Y, Genome Instability and Aging. Annual Review of Physiology, 2013. 75(1): p. 645–668. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

