Telomere-to-telomere assembly of the genome of an individual Oikopleura dioica from Okinawa using Nanopore-based sequencing
- PMID: 33781200
- PMCID: PMC8008620
- DOI: 10.1186/s12864-021-07512-6
Telomere-to-telomere assembly of the genome of an individual Oikopleura dioica from Okinawa using Nanopore-based sequencing
Abstract
Background: The larvacean Oikopleura dioica is an abundant tunicate plankton with the smallest (65-70 Mbp) non-parasitic, non-extremophile animal genome identified to date. Currently, there are two genomes available for the Bergen (OdB3) and Osaka (OSKA2016) O. dioica laboratory strains. Both assemblies have full genome coverage and high sequence accuracy. However, a chromosome-scale assembly has not yet been achieved.
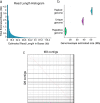
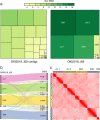
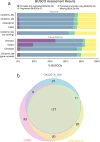
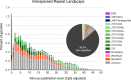
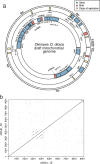
Results: Here, we present a chromosome-scale genome assembly (OKI2018_I69) of the Okinawan O. dioica produced using long-read Nanopore and short-read Illumina sequencing data from a single male, combined with Hi-C chromosomal conformation capture data for scaffolding. The OKI2018_I69 assembly has a total length of 64.3 Mbp distributed among 19 scaffolds. 99% of the assembly is contained within five megabase-scale scaffolds. We found telomeres on both ends of the two largest scaffolds, which represent assemblies of two fully contiguous autosomal chromosomes. Each of the other three large scaffolds have telomeres at one end only and we propose that they correspond to sex chromosomes split into a pseudo-autosomal region and X-specific or Y-specific regions. Indeed, these five scaffolds mostly correspond to equivalent linkage groups in OdB3, suggesting overall agreement in chromosomal organization between the two populations. At a more detailed level, the OKI2018_I69 assembly possesses similar genomic features in gene content and repetitive elements reported for OdB3. The Hi-C map suggests few reciprocal interactions between chromosome arms. At the sequence level, multiple genomic features such as GC content and repetitive elements are distributed differently along the short and long arms of the same chromosome.
Conclusions: We show that a hybrid approach of integrating multiple sequencing technologies with chromosome conformation information results in an accurate de novo chromosome-scale assembly of O. dioica's highly polymorphic genome. This genome assembly opens up the possibility of cross-genome comparison between O. dioica populations, as well as of studies of chromosomal evolution in this lineage.
Keywords: Chromosome-scale assembly; Hi-C; Oikopleura dioica; Oxford Nanopore sequencing; Single individual; Telomere-to-telomere.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Alldredge AL. Discarded appendicularian houses as sources of food, surface habitats, and particulate organic matter in planktonic environments. Limnol Oceanogr. 1976;21(1):14–24. doi: 10.4319/lo.1976.21.1.0014. - DOI
-
- Hopcroft RR, Roff JC. Zooplankton growth rates: extraordinary production by the larvacean Oikopleura dioica in tropical waters. J Plankton Res. 1995;17(2):205–220. doi: 10.1093/plankt/17.2.205. - DOI
-
- Sato R, Tanaka Y, Ishimaru T. House production by Oikopleura dioica (Tunicata, Appendicularia) under laboratory conditions. J Plankton Res. 2001;23(4):415–423. doi: 10.1093/plankt/23.4.415. - DOI
-
- Alldredge A. The contribution of discarded appendicularian houses to the flux of particulate organic carbon from oceanic surface waters. In: Gorsky G, Youngbluth MJ, Deibel D, editors. Response of Marine Ecosystems to Global Change: Ecological Impact of Appendicularians: Contemporaty Publishing International; 2005. p. 309–26.
-
- Masunaga A, Liu AW, Tan Y, Scott A, Luscombe NM. Streamlined sampling and cultivation of the pelagic cosmopolitan larvacean, Oikopleura dioica. JoVE (Journal of Visualized Experiments). 2020;16(160):e61279. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

