Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium
- PMID: 33781205
- PMCID: PMC8007386
- DOI: 10.1186/s12864-021-07528-y
Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium
Abstract
Background: Understanding viral infection of the olfactory epithelium is essential because the olfactory nerve is an important route of entry for viruses to the central nervous system. Specialized chemosensory epithelial cells that express the transient receptor potential cation channel subfamily M member 5 (TRPM5) are found throughout the airways and intestinal epithelium and are involved in responses to viral infection.
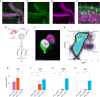
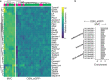
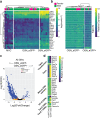
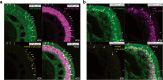
Results: Herein we performed deep transcriptional profiling of olfactory epithelial cells sorted by flow cytometry based on the expression of mCherry as a marker for olfactory sensory neurons and for eGFP in OMP-H2B::mCherry/TRPM5-eGFP transgenic mice (Mus musculus). We find profuse expression of transcripts involved in inflammation, immunity and viral infection in TRPM5-expressing microvillous cells compared to olfactory sensory neurons.
Conclusion: Our study provides new insights into a potential role for TRPM5-expressing microvillous cells in viral infection of the olfactory epithelium. We find that, as found for solitary chemosensory cells (SCCs) and brush cells in the airway epithelium, and for tuft cells in the intestine, the transcriptome of TRPM5-expressing microvillous cells indicates that they are likely involved in the inflammatory response elicited by viral infection of the olfactory epithelium.
Keywords: Immunity; Inflammation; Microvillous cells; Mouse; Olfactory sensory neurons; Viral infection.
Conflict of interest statement
The authors declare no competing interests.
Figures

Update of
-
Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium.bioRxiv [Preprint]. 2020 Dec 1:2020.05.14.096016. doi: 10.1101/2020.05.14.096016. bioRxiv. 2020. Update in: BMC Genomics. 2021 Mar 30;22(1):224. doi: 10.1186/s12864-021-07528-y. PMID: 32511400 Free PMC article. Updated. Preprint.
Similar articles
-
Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium.bioRxiv [Preprint]. 2020 Dec 1:2020.05.14.096016. doi: 10.1101/2020.05.14.096016. bioRxiv. 2020. Update in: BMC Genomics. 2021 Mar 30;22(1):224. doi: 10.1186/s12864-021-07528-y. PMID: 32511400 Free PMC article. Updated. Preprint.
-
Skn-1a/Pou2f3 is required for the generation of Trpm5-expressing microvillous cells in the mouse main olfactory epithelium.BMC Neurosci. 2014 Jan 16;15:13. doi: 10.1186/1471-2202-15-13. BMC Neurosci. 2014. PMID: 24428937 Free PMC article.
-
Is TrpM5 a reliable marker for chemosensory cells? Multiple types of microvillous cells in the main olfactory epithelium of mice.BMC Neurosci. 2008 Dec 4;9:115. doi: 10.1186/1471-2202-9-115. BMC Neurosci. 2008. PMID: 19055837 Free PMC article.
-
TRPM5.Handb Exp Pharmacol. 2014;222:489-502. doi: 10.1007/978-3-642-54215-2_19. Handb Exp Pharmacol. 2014. PMID: 24756718 Review.
-
TRPM5 in the battle against diabetes and obesity.Acta Physiol (Oxf). 2018 Feb;222(2). doi: 10.1111/apha.12949. Epub 2017 Oct 4. Acta Physiol (Oxf). 2018. PMID: 28834354 Review.
Cited by
-
Modulation of olfactory signal detection in the olfactory epithelium: focus on the internal and external environment, and the emerging role of the immune system.Cell Tissue Res. 2021 Jun;384(3):589-605. doi: 10.1007/s00441-021-03467-y. Epub 2021 May 7. Cell Tissue Res. 2021. PMID: 33961125 Free PMC article. Review.
-
SARS-COV-2, infection, transmission, transcription, translation, proteins, and treatment: A review.Int J Biol Macromol. 2021 Dec 15;193(Pt B):1249-1273. doi: 10.1016/j.ijbiomac.2021.10.172. Epub 2021 Oct 28. Int J Biol Macromol. 2021. PMID: 34756970 Free PMC article. Review.
-
Neuroimmune interactions in the olfactory epithelium: maintaining a sensory organ at an immune barrier interface.Trends Immunol. 2024 Dec;45(12):987-1000. doi: 10.1016/j.it.2024.10.005. Epub 2024 Nov 15. Trends Immunol. 2024. PMID: 39550314 Review.
-
Identification and profiling of Trichinella spiralis circulating antigens and proteins in sera of mice with trichinellosis.PLoS One. 2022 Mar 10;17(3):e0265013. doi: 10.1371/journal.pone.0265013. eCollection 2022. PLoS One. 2022. PMID: 35271623 Free PMC article.
-
The olfactory nerve is not a likely route to brain infection in COVID-19: a critical review of data from humans and animal models.Acta Neuropathol. 2021 Jun;141(6):809-822. doi: 10.1007/s00401-021-02314-2. Epub 2021 Apr 26. Acta Neuropathol. 2021. PMID: 33903954 Free PMC article. Review.
References
-
- Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology (R Package) 2020.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

