ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis
- PMID: 33782125
- PMCID: PMC8040812
- DOI: 10.1073/pnas.2021671118
ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis
Abstract
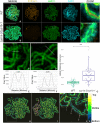
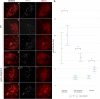
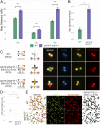
The synaptonemal complex is a tripartite proteinaceous ultrastructure that forms between homologous chromosomes during prophase I of meiosis in the majority of eukaryotes. It is characterized by the coordinated installation of transverse filament proteins between two lateral elements and is required for wild-type levels of crossing over and meiotic progression. We have generated null mutants of the duplicated Arabidopsis transverse filament genes zyp1a and zyp1b using a combination of T-DNA insertional mutants and targeted CRISPR/Cas mutagenesis. Cytological and genetic analysis of the zyp1 null mutants reveals loss of the obligate chiasma, an increase in recombination map length by 1.3- to 1.7-fold and a virtual absence of cross-over (CO) interference, determined by a significant increase in the number of double COs. At diplotene, the numbers of HEI10 foci, a marker for Class I interference-sensitive COs, are twofold greater in the zyp1 mutant compared to wild type. The increase in recombination in zyp1 does not appear to be due to the Class II interference-insensitive COs as chiasmata were reduced by ∼52% in msh5/zyp1 compared to msh5 These data suggest that ZYP1 limits the formation of closely spaced Class I COs in Arabidopsis Our data indicate that installation of ZYP1 occurs at ASY1-labeled axial bridges and that loss of the protein disrupts progressive coalignment of the chromosome axes.
Keywords: cross-over interference; meiosis; recombination; synaptonemal complex.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures




Comment in
References
-
- Zickler D., Kleckner N., Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33, 603–754 (1999). - PubMed
-
- Page S. L., Hawley R. S., The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20, 525–558 (2004). - PubMed
-
- Kleckner N., Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

