Pharyngeal Microbial Signatures Are Predictive of the Risk of Fungal Pneumonia in Hematologic Patients
- PMID: 33782152
- PMCID: PMC8281212
- DOI: 10.1128/IAI.00105-21
Pharyngeal Microbial Signatures Are Predictive of the Risk of Fungal Pneumonia in Hematologic Patients
Abstract
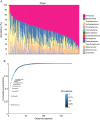
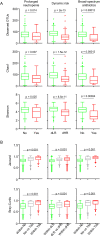
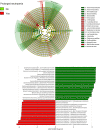
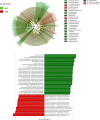
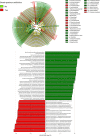
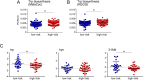
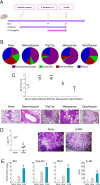
The ability to predict invasive fungal infections (IFI) in patients with hematological malignancies is fundamental for successful therapy. Although gut dysbiosis is known to occur in hematological patients, whether airway dysbiosis also contributes to the risk of IFI has not been investigated. Nasal and oropharyngeal swabs were collected for functional microbiota characterization in 173 patients with hematological malignancies recruited in a multicenter, prospective, observational study and stratified according to the risk of developing IFI. A lower microbial richness and evenness were found in the pharyngeal microbiota of high-risk patients that were associated with a distinct taxonomic and metabolic profile. A murine model of IFI provided biologic plausibility for the finding that loss of protective anaerobes, such as Clostridiales and Bacteroidetes, along with an apparent restricted availability of tryptophan, is causally linked to the risk of IFI in hematologic patients and indicates avenues for antimicrobial stewardship and metabolic reequilibrium in IFI.
Keywords: airway microbiome; antibiotics; hematological malignancies; indole-3-aldehyde; invasive fungal infection; metabolomics; tryptophan.
Figures
Comment in
- Infect Immun. 89.
References
-
- Ananda-Rajah MR, Cheng A, Morrissey CO, Spelman T, Dooley M, Neville AM, Slavin M. 2011. Attributable hospital cost and antifungal treatment of invasive fungal diseases in high-risk hematology patients: an economic modeling approach. Antimicrob Agents Chemother 55:1953–1960. 10.1128/AAC.01423-10. - DOI - PMC - PubMed
-
- Pagano L, Busca A, Candoni A, Cattaneo C, Cesaro S, Fanci R, Nadali G, Potenza L, Russo D, Tumbarello M, Nosari A, Aversa F, et al. 2017. Risk stratification for invasive fungal infections in patients with hematological malignancies: SEIFEM recommendations. Blood Rev 31:17–29. 10.1016/j.blre.2016.09.002. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

