MiR-199-3p enhances muscle regeneration and ameliorates aged muscle and muscular dystrophy
- PMID: 33782502
- PMCID: PMC8007565
- DOI: 10.1038/s42003-021-01952-2
MiR-199-3p enhances muscle regeneration and ameliorates aged muscle and muscular dystrophy
Abstract
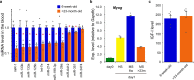
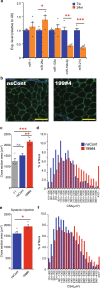
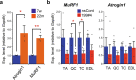
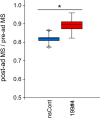
Parabiosis experiments suggest that molecular factors related to rejuvenation and aging circulate in the blood. Here, we show that miR-199-3p, which circulates in the blood as a cell-free miRNA, is significantly decreased in the blood of aged mice compared to young mice; and miR-199-3p has the ability to enhance myogenic differentiation and muscle regeneration. Administration of miR-199 mimics, which supply miR-199-3p, to aged mice resulted in muscle fiber hypertrophy and delayed loss of muscle strength. Systemic administration of miR-199 mimics to mdx mice, a well-known animal model of Duchenne muscular dystrophy (DMD), markedly improved the muscle strength of mice. Taken together, cell-free miR-199-3p in the blood may have an anti-aging effect such as a hypertrophic effect in aged muscle fibers and could have potential as a novel RNA therapeutic for DMD as well as age-related diseases. The findings provide us with new insights into blood-circulating miRNAs as age-related molecules.
Conflict of interest statement
H.H. and M.F. have a pending patent regarding the results of this study. All other authors declare no competing interests.
Figures




Similar articles
-
Alterations in Notch signalling in skeletal muscles from mdx and dko dystrophic mice and patients with Duchenne muscular dystrophy.Exp Physiol. 2014 Apr;99(4):675-87. doi: 10.1113/expphysiol.2013.077255. Epub 2014 Jan 17. Exp Physiol. 2014. PMID: 24443351
-
miR-146a deficiency does not aggravate muscular dystrophy in mdx mice.Skelet Muscle. 2019 Aug 14;9(1):22. doi: 10.1186/s13395-019-0207-0. Skelet Muscle. 2019. PMID: 31412923 Free PMC article.
-
MicroRNA-33 inhibition ameliorates muscular dystrophy by enhancing skeletal muscle regeneration.EMBO Mol Med. 2025 Aug;17(8):1902-1925. doi: 10.1038/s44321-025-00273-9. Epub 2025 Jul 23. EMBO Mol Med. 2025. PMID: 40695994 Free PMC article.
-
Functional characteristics of dystrophic skeletal muscle: insights from animal models.J Appl Physiol (1985). 2002 Aug;93(2):407-17. doi: 10.1152/japplphysiol.01242.2001. J Appl Physiol (1985). 2002. PMID: 12133845 Review.
-
Markers of oxidative stress and aging in Duchene muscular dystrophy patients and the possible ameliorating effect of He:Ne laser.Acta Myol. 2007 Jul;26(1):14-21. Acta Myol. 2007. PMID: 17915565 Free PMC article. Review.
Cited by
-
Saikosaponin D alleviates inflammatory response of osteoarthritis and mediates autophagy via elevating microRNA-199-3p to target transcription Factor-4.J Orthop Surg Res. 2024 Feb 22;19(1):151. doi: 10.1186/s13018-024-04607-0. J Orthop Surg Res. 2024. PMID: 38389105 Free PMC article.
-
The Role of the Skeletal Muscle Secretome in Mediating Endurance and Resistance Training Adaptations.Front Physiol. 2021 Aug 12;12:709807. doi: 10.3389/fphys.2021.709807. eCollection 2021. Front Physiol. 2021. PMID: 34456749 Free PMC article. Review.
-
Dynamic Aging: Channeled Through Microenvironment.Front Physiol. 2021 Jul 21;12:702276. doi: 10.3389/fphys.2021.702276. eCollection 2021. Front Physiol. 2021. PMID: 34366891 Free PMC article. Review.
-
Therapies Targeted at Non-Coding RNAs in Prevention and Limitation of Myocardial Infarction and Subsequent Cardiac Remodeling-Current Experience and Perspectives.Int J Mol Sci. 2021 May 27;22(11):5718. doi: 10.3390/ijms22115718. Int J Mol Sci. 2021. PMID: 34071976 Free PMC article. Review.
-
Droplet Digital PCR Quantification of Selected Intracellular and Extracellular microRNAs Reveals Changes in Their Expression Pattern during Porcine In Vitro Adipogenesis.Genes (Basel). 2023 Mar 9;14(3):683. doi: 10.3390/genes14030683. Genes (Basel). 2023. PMID: 36980955 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

