Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries
- PMID: 33782558
- PMCID: PMC8007471
- DOI: 10.1038/s41564-021-00870-7
Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries
Abstract
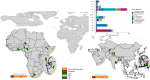
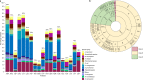
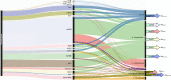
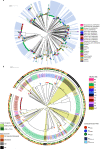
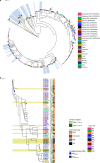
Antimicrobial resistance in neonatal sepsis is rising, yet mechanisms of resistance that often spread between species via mobile genetic elements, ultimately limiting treatments in low- and middle-income countries (LMICs), are poorly characterized. The Burden of Antibiotic Resistance in Neonates from Developing Societies (BARNARDS) network was initiated to characterize the cause and burden of antimicrobial resistance in neonatal sepsis for seven LMICs in Africa and South Asia. A total of 36,285 neonates were enrolled in the BARNARDS study between November 2015 and December 2017, of whom 2,483 were diagnosed with culture-confirmed sepsis. Klebsiella pneumoniae (n = 258) was the main cause of neonatal sepsis, with Serratia marcescens (n = 151), Klebsiella michiganensis (n = 117), Escherichia coli (n = 75) and Enterobacter cloacae complex (n = 57) also detected. We present whole-genome sequencing, antimicrobial susceptibility and clinical data for 916 out of 1,038 neonatal sepsis isolates (97 isolates were not recovered from initial isolation at local sites). Enterobacterales (K. pneumoniae, E. coli and E. cloacae) harboured multiple cephalosporin and carbapenem resistance genes. All isolated pathogens were resistant to multiple antibiotic classes, including those used to treat neonatal sepsis. Intraspecies diversity of K. pneumoniae and E. coli indicated that multiple antibiotic-resistant lineages cause neonatal sepsis. Our results will underpin research towards better treatments for neonatal sepsis in LMICs.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- World Mortality 2019 (Department of Economic and Social Affairs, United Nations, 2019).
-
- Shukla V, Mwenechanya M, Carlo WA. Dealing with neonatal emergencies in low-resource settings. Semin. Fetal Neonatal Med. 2019;24:101028. - PubMed
-
- Popescu CR, et al. Neonatal sepsis in low-income countries: epidemiology, diagnosis and prevention. Expert Rev. Anti. Infect. Ther. 2020;18:443–452. - PubMed
-
- Folgori L, Bielicki J, Heath PT, Sharland M. Antimicrobial-resistant Gram-negative infections in neonates: burden of disease and challenges in treatment. Curr. Opin. Infect. Dis. 2017;30:281–288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

