Ultraconserved enhancer function does not require perfect sequence conservation
- PMID: 33782603
- PMCID: PMC8038972
- DOI: 10.1038/s41588-021-00812-3
Ultraconserved enhancer function does not require perfect sequence conservation
Abstract
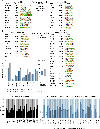
Ultraconserved enhancer sequences show perfect conservation between human and rodent genomes, suggesting that their functions are highly sensitive to mutation. However, current models of enhancer function do not sufficiently explain this extreme evolutionary constraint. We subjected 23 ultraconserved enhancers to different levels of mutagenesis, collectively introducing 1,547 mutations, and examined their activities in transgenic mouse reporter assays. Overall, we find that the regulatory properties of ultraconserved enhancers are robust to mutation. Upon mutagenesis, nearly all (19/23, 83%) still functioned as enhancers at one developmental stage, as did most of those tested again later in development (5/9, 56%). Replacement of endogenous enhancers with mutated alleles in mice corroborated results of transgenic assays, including the functional resilience of ultraconserved enhancers to mutation. Our findings show that the currently known activities of ultraconserved enhancers do not necessarily require the perfect conservation observed in evolution and suggest that additional regulatory or other functions contribute to their sequence constraint.
Conflict of interest statement
Competing Interest Statements
J.L.R.R. is cofounder, stockholder, and currently on the scientific board of Neurona, a company studying the potential therapeutic use of interneuron transplantation. The authors declare no other competing interests.
Figures













Comment in
-
Ultraconservation of enhancers is not ultranecessary.Nat Genet. 2021 Apr;53(4):429-430. doi: 10.1038/s41588-021-00839-6. Nat Genet. 2021. PMID: 33782604 No abstract available.
References
-
- Bejerano G et al. Ultraconserved elements in the human genome. Science 304, 1321–5 (2004). - PubMed
-
- Katzman S et al. Human genome ultraconserved elements are ultraselected. Science 317, 915 (2007). - PubMed
-
- Drake JA et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet 38, 223–7 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

