TRIM28 functions as a negative regulator of aggresome formation
- PMID: 33783327
- PMCID: PMC8726693
- DOI: 10.1080/15548627.2021.1909835
TRIM28 functions as a negative regulator of aggresome formation
Abstract
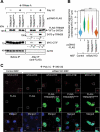
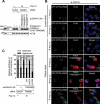
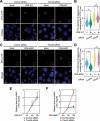
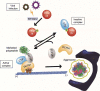
Selective recognition and elimination of misfolded polypeptides are crucial for protein homeostasis. When the ubiquitin-proteasome system is impaired, misfolded polypeptides tend to form small cytosolic aggregates and are transported to the aggresome and eventually eliminated by the autophagy pathway. Despite the importance of this process, the regulation of aggresome formation remains poorly understood. Here, we identify TRIM28/TIF1β/KAP1 (tripartite motif containing 28) as a negative regulator of aggresome formation. Direct interaction between TRIM28 and CTIF (cap binding complex dependent translation initiation factor) leads to inefficient aggresomal targeting of misfolded polypeptides. We also find that either treatment of cells with poly I:C or infection of the cells by influenza A viruses triggers the phosphorylation of TRIM28 at S473 in a way that depends on double-stranded RNA-activated protein kinase. The phosphorylation promotes association of TRIM28 with CTIF, inhibits aggresome formation, and consequently suppresses viral proliferation. Collectively, our data provide compelling evidence that TRIM28 is a negative regulator of aggresome formation.Abbreviations: BAG3: BCL2-associated athanogene 3; CTIF: CBC-dependent translation initiation factor; CED: CTIF-EEF1A1-DCTN1; DCTN1: dynactin subunit 1; EEF1A1: eukaryotic translation elongation factor 1 alpha 1; EIF2AK2: eukaryotic translation initiation factor 2 alpha kinase 2; HDAC6: histone deacetylase 6; IAV: influenza A virus; IP: immunoprecipitation; PLA: proximity ligation assay; polypeptidyl-puro: polypeptidyl-puromycin; qRT-PCR: quantitative reverse-transcription PCR; siRNA: small interfering RNA.
Keywords: Aggrephagy; CTIF; DCTN1; EEF1A1; EIF2AK2; influenza A virus.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
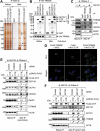
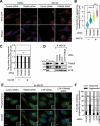
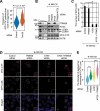
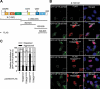
References
-
- Caughey B, Lansbury PT.. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. - PubMed
-
- Chiti F, Dobson CM.. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
-
- Richter-Landsberg C, Leyk J. Inclusion body formation, macroautophagy, and the role of HDAC6 in neurodegeneration. Acta Neuropathol. 2013;126:793–807. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
