Sizing Extracellular Vesicles Using Membrane Dyes and a Single Molecule-Sensitive Flow Analyzer
- PMID: 33784071
- PMCID: PMC10243643
- DOI: 10.1021/acs.analchem.1c00253
Sizing Extracellular Vesicles Using Membrane Dyes and a Single Molecule-Sensitive Flow Analyzer
Abstract
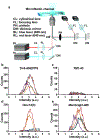
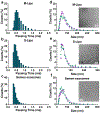
Extracellular vesicles (EVs) are membranous particles released by most cells in our body, which are involved in many cell-to-cell signaling processes. Given the nanometer sizes and heterogeneity of EVs, highly sensitive methods with single-molecule resolution are fundamental to investigating their biophysical properties. Here, we demonstrate the sizing of EVs using a fluorescence-based flow analyzer with single-molecule sensitivity. Using a dye that selectively partitions into the vesicle's membrane, we show that the fluorescence intensity of a vesicle is proportional to its diameter. We discuss the constraints in sample preparation which are inherent to sizing nanoscale vesicles with a fluorescent membrane dye and propose several guidelines to improve data consistency. After optimizing staining conditions, we were able to measure the size of vesicles in the range ∼35-300 nm, covering the spectrum of EV sizes. Lastly, we developed a method to correct the signal intensity from each vesicle based on its traveling speed inside the microfluidic channel, by operating at a high sampling rate (10 kHz) and measuring the time required for the particle to cross the laser beam. Using this correction, we obtained a threefold greater accuracy in EV sizing, with a precision of ±15-25%.
Figures


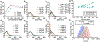
References
-
- Greening DW; Xu R; Ji H; Tauro BJ; Simpson RJ (2015) A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. In: Posch A (eds) Proteomic Profiling. Methods in Molecular Biology, vol 1295. Humana Press, New York, NY. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

