HIV-1 integration sites in CD4+ T cells during primary, chronic, and late presentation of HIV-1 infection
- PMID: 33784259
- PMCID: PMC8262285
- DOI: 10.1172/jci.insight.143940
HIV-1 integration sites in CD4+ T cells during primary, chronic, and late presentation of HIV-1 infection
Abstract
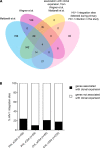
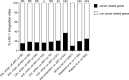
HIV-1 is capable of integrating its genome into that of its host cell. We examined the influence of the activation state of CD4+ T cells, the effect of antiretroviral therapy (ART), and the clinical stage of HIV-1 infection on HIV-1 integration site features and selection. HIV-1 integration sites were sequenced from longitudinally sampled resting and activated CD4+ T cells from 12 HIV-1-infected individuals. In total, 589 unique HIV-1 integration sites were analyzed: 147, 391, and 51 during primary, chronic, and late presentation of HIV-1 infection, respectively. As early as during primary HIV-1 infection and independent of the activation state of CD4+ T cells collected on and off ART, HIV-1 integration sites were preferentially detected in recurrent integration genes, genes associated with clonal expansion of latently HIV-1-infected CD4+ T cells, cancer-related genes, and highly expressed genes. The preference for cancer-related genes was more pronounced at late stages of HIV-1 infection. Host genomic features of HIV-1 integration site selection remained stable during HIV-1 infection in both resting and activated CD4+ T cells. In summary, characteristic HIV-1 integration site features are preestablished as early as during primary HIV-1 infection and are found in both resting and activated CD4+ T cells.
Keywords: AIDS/HIV; Molecular biology; T cells; Virology.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

