POT1 stability and binding measured by fluorescence thermal shift assays
- PMID: 33784306
- PMCID: PMC8009405
- DOI: 10.1371/journal.pone.0245675
POT1 stability and binding measured by fluorescence thermal shift assays
Abstract
The protein POT1 (Protection of Telomeres 1) is an integral part of the shelterin complex that protects the ends of human chromosomes from degradation or end fusions. It is the only component of shelterin that binds single-stranded DNA. We describe here the application of two separate fluorescent thermal shift assays (FTSA) that provide quantitative biophysical characterization of POT1 stability and its interactions. The first assay uses Sypro Orange™ and monitors the thermal stability of POT1 and its binding under a variety of conditions. This assay is useful for the quality control of POT1 preparations, for biophysical characterization of its DNA binding and, potentially, as an efficient screening tool for binding of small molecule drug candidates. The second assay uses a FRET-labeled human telomeric G-quadruplex structure that reveals the effects of POT1 binding on thermal stability from the DNA frame of reference. These complementary assays provide efficient biophysical approaches for the quantitative characterization of multiple aspects of POT1 structure and function. The results from these assays provide thermodynamics details of POT1 folding, the sequence selectivity of its DNA binding and the thermodynamic profile for its binding to its preferred DNA binding sequence. Most significantly, results from these assays elucidate two mechanisms for the inhibition of POT1 -DNA interactions. The first is by competitive inhibition at the POT1 DNA binding site. The second is indirect and is by stabilization of G-quadruplex formation within the normal POT1 single-stranded DNA sequence to prevent POT1 binding.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
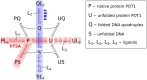


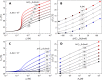

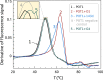
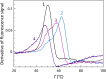


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

