A Recombinant Fragment of Human Surfactant Protein D Binds Spike Protein and Inhibits Infectivity and Replication of SARS-CoV-2 in Clinical Samples
- PMID: 33784482
- PMCID: PMC8320127
- DOI: 10.1165/rcmb.2021-0005OC
A Recombinant Fragment of Human Surfactant Protein D Binds Spike Protein and Inhibits Infectivity and Replication of SARS-CoV-2 in Clinical Samples
Abstract
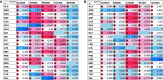
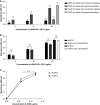
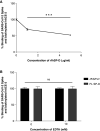
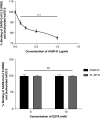
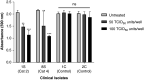
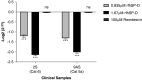
Coronavirus disease (COVID-19) is an acute infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Human SP-D (surfactant protein D) is known to interact with the spike protein of SARS-CoV, but its immune surveillance against SARS-CoV-2 is not known. The current study aimed to examine the potential of a recombinant fragment of human SP-D (rfhSP-D) as an inhibitor of replication and infection of SARS-CoV-2. The interaction of rfhSP-D with the spike protein of SARS-CoV-2 and human ACE-2 (angiotensin-converting enzyme 2) receptor was predicted via docking analysis. The inhibition of interaction between the spike protein and ACE-2 by rfhSP-D was confirmed using direct and indirect ELISA. The effect of rfhSP-D on replication and infectivity of SARS-CoV-2 from clinical samples was assessed by measuring the expression of RdRp gene of the virus using quantitative PCR. In silico interaction studies indicated that three amino acid residues in the receptor-binding domain of spike protein of SARS-CoV-2 were commonly involved in interacting with rfhSP-D and ACE-2. Studies using clinical samples of SARS-CoV-2-positive cases (asymptomatic, n = 7; symptomatic, n = 8) and negative control samples (n = 15) demonstrated that treatment with 1.67 μM rfhSP-D inhibited viral replication by ∼5.5-fold and was more efficient than remdesivir (100 μM) in Vero cells. An approximately two-fold reduction in viral infectivity was also observed after treatment with 1.67 μM rfhSP-D. These results conclusively demonstrate that the rfhSP-D mediated calcium independent interaction between the receptor-binding domain of the S1 subunit of the SARS-CoV-2 spike protein and human ACE-2, its host cell receptor, and significantly reduced SARS-CoV-2 infection and replication in vitro.
Keywords: COVID-19; SARS-CoV-2; entry inhibitor; spike protein; surfactant protein D.
Figures



References
-
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. - PMC - PubMed
-
- World Health Organization. Geneva, Switzerland: World Health Organization; 2020. https://www.who.int/publications/m/item/weekly-epidemiological-update---...
-
- National Institutes of Health. Bethesda, MD: National Institutes of Health; https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

