Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication
- PMID: 33784499
- PMCID: PMC7969873
- DOI: 10.1016/j.celrep.2021.108940
Antiviral drug screen identifies DNA-damage response inhibitor as potent blocker of SARS-CoV-2 replication
Abstract
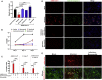
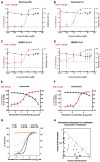
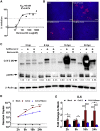
SARS-CoV-2 has currently precipitated the COVID-19 global health crisis. We developed a medium-throughput drug-screening system and identified a small-molecule library of 34 of 430 protein kinase inhibitors that were capable of inhibiting the SARS-CoV-2 cytopathic effect in human epithelial cells. These drug inhibitors are in various stages of clinical trials. We detected key proteins involved in cellular signaling pathways mTOR-PI3K-AKT, ABL-BCR/MAPK, and DNA-damage response that are critical for SARS-CoV-2 infection. A drug-protein interaction-based secondary screen confirmed compounds, such as the ATR kinase inhibitor berzosertib and torin2 with anti-SARS-CoV-2 activity. Berzosertib exhibited potent antiviral activity against SARS-CoV-2 in multiple cell types and blocked replication at the post-entry step. Berzosertib inhibited replication of SARS-CoV-1 and the Middle East respiratory syndrome coronavirus (MERS-CoV) as well. Our study highlights key promising kinase inhibitors to constrain coronavirus replication as a host-directed therapy in the treatment of COVID-19 and beyond as well as provides an important mechanism of host-pathogen interactions.
Keywords: ATR kinase; COVID-19; DNA-damage response pathway; SARS-CoV-2; berzosertib; high-throughput screen; mTOR-PI3K-AKT pathway; nucleoside analogs; protein kinase inhibitors.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests U.A.K.B. is an employee of Merck KGaA, Darmstadt, Germany. Berzosertib compound is licensed by Merck KGaA, Darmstadt, Germany. The other authors declare no competing financial interests.
Figures

References
-
- Belhadjer Z., Méot M., Bajolle F., Khraiche D., Legendre A., Abakka S., Auriau J., Grimaud M., Oualha M., Beghetti M. Acute heart failure in multisystem inflammatory syndrome in children (MIS-C) in the context of global SARS-CoV-2 pandemic. Circulation. 2020;142:429–436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

