Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox
- PMID: 33785070
- PMCID: PMC8008609
- DOI: 10.1186/s13059-021-02306-1
Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox
Abstract
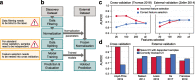
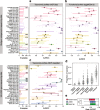
The human microbiome is increasingly mined for diagnostic and therapeutic biomarkers using machine learning (ML). However, metagenomics-specific software is scarce, and overoptimistic evaluation and limited cross-study generalization are prevailing issues. To address these, we developed SIAMCAT, a versatile R toolbox for ML-based comparative metagenomics. We demonstrate its capabilities in a meta-analysis of fecal metagenomic studies (10,803 samples). When naively transferred across studies, ML models lost accuracy and disease specificity, which could however be resolved by a novel training set augmentation strategy. This reveals some biomarkers to be disease-specific, with others shared across multiple conditions. SIAMCAT is freely available from siamcat.embl.de .
Keywords: Machine learning; Meta-analysis; Microbiome data analysis; Microbiome-wide association studies (MWAS); Statistical modeling.
Conflict of interest statement
The authors declared that they have no competing interests.
Figures




References
-
- Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. Mass Medical Soc; 2016;375:2369–2379. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

