The global dissemination of hospital clones of Enterococcus faecium
- PMID: 33785076
- PMCID: PMC8008517
- DOI: 10.1186/s13073-021-00868-0
The global dissemination of hospital clones of Enterococcus faecium
Abstract
Background: The hospital-adapted A1 group of Enterococcus faecium remains an organism of significant concern in the context of drug-resistant hospital-associated infections. How this pathogen evolves and disseminates remains poorly understood.
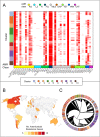
Methods: A large, globally representative collection of short-read genomic data from the hospital-associated A1 group of Enterococcus faecium was assembled (n = 973). We analysed, using a novel analysis approach, global diversity in terms of both the dynamics of the accessory genome and homologous recombination among conserved genes.
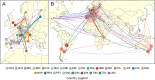
Results: Two main modes of genomic evolution continue to shape E. faecium: the acquisition and loss of genes, including antimicrobial resistance genes, through mobile genetic elements including plasmids, and homologous recombination of the core genome. These events lead to new clones emerging at the local level, followed by the erosion of signals of clonality through recombination, and in some identifiable cases producing new clonal clusters. These patterns lead to new, emerging lineages which are able to spread globally over relatively short timeframes.
Conclusions: The ability of A1 E. faecium to continually present new combinations of genes for potential selection suggests that controlling this pathogen will remain challenging but establishing a framework for understanding genomic evolution is likely to aid in tracking the threats posed by newly emerging lineages.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011-2014. Infect Control Hosp Epidemiol. 2016;37(11):1288–1301. doi: 10.1017/ice.2016.174. - DOI - PMC - PubMed
-
- van Hal SJ, Beukers AG, Timms VJ, Ellem JA, Taylor P, Maley MW, Newton PJ, Ferguson JK, Lee A, Chen SC, Sintchenko V. Relentless spread and adaptation of non-typeable vanA vancomycin-resistant Enterococcus faecium: a genome-wide investigation. J Antimicrob Chemother. 2018;73(6):1487–1491. doi: 10.1093/jac/dky074. - DOI - PubMed
-
- Ayobami O, Willrich N, Reuss A, Eckmanns T, Markwart R. The ongoing challenge of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis in Europe: an epidemiological analysis of bloodstream infections. Emerg Microbes Infect. 2020;9(1):1180–1193. doi: 10.1080/22221751.2020.1769500. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

