Emergence and outcomes of the SARS-CoV-2 'Marseille-4' variant
- PMID: 33785459
- PMCID: PMC7997945
- DOI: 10.1016/j.ijid.2021.03.068
Emergence and outcomes of the SARS-CoV-2 'Marseille-4' variant
Abstract
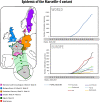
Background: In Marseille, France, following a first severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak in March-May 2020, a second epidemic phase occurred from June, involving 10 new variants. The Marseille-4 variant caused an epidemic that started in August and is still ongoing.
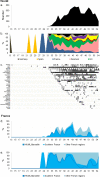
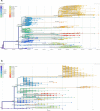
Methods: The 1038 SARS-CoV-2 whole genome sequences obtained in our laboratory by next-generation sequencing with Illumina technology were analysed using Nextclade and nextstrain/ncov pipelines and IQ-TREE. A Marseille-4-specific qPCR assay was implemented. Demographic and clinical features were compared between patients with the Marseille-4 variant and those with earlier strains.
Results: Marseille-4 harbours 13 hallmark mutations. One leads to an S477N substitution in the receptor binding domain of the spike protein targeted by current vaccines. Using a specific qPCR, it was observed that Marseille-4 caused 12-100% of SARS-CoV-2 infections in Marseille from September 2020, being involved in 2106 diagnoses. This variant was more frequently associated with hypoxemia than were clade 20A strains before May 2020. It caused a re-infection in 11 patients diagnosed with different SARS-CoV-2 strains before June 2020, suggesting either short-term protective immunity or a lack of cross-immunity.
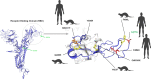
Conclusions: Marseille-4 should be considered as a major SARS-CoV-2 variant. Its sudden appearance points towards an animal reservoir, possibly mink. The protective role of past exposure and current vaccines against this variant should be evaluated.
Keywords: COVID-19; Marseille-4; Molecular epidemiology; Mutations; SARS-CoV-2; Spike; Variant.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

