Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2
- PMID: 33785622
- PMCID: PMC8092272
- DOI: 10.1128/mBio.02924-20
Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2
Abstract
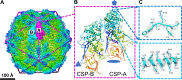
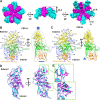
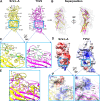
Trichomonas vaginalis, the causative pathogen for the most common nonviral sexually transmitted infection worldwide, is itself frequently infected with one or more of the four types of small double-stranded RNA (dsRNA) Trichomonas vaginalis viruses (TVV1 to 4, genus Trichomonasvirus, family Totiviridae). Each TVV encloses a nonsegmented genome within a single-layered capsid and replicates entirely intracellularly, like many dsRNA viruses, and unlike those in the Reoviridae family. Here, we have determined the structure of TVV2 by cryo-electron microscopy (cryoEM) at 3.6 Å resolution and derived an atomic model of its capsid. TVV2 has an icosahedral, T = 2*, capsid comprised of 60 copies of the icosahedral asymmetric unit (a dimer of the two capsid shell protein [CSP] conformers, CSP-A and CSP-B), typical of icosahedral dsRNA virus capsids. However, unlike the robust CSP-interlocking interactions such as the use of auxiliary "clamping" proteins among Reoviridae, only lateral CSP interactions are observed in TVV2, consistent with an assembly strategy optimized for TVVs' intracellular-only replication cycles within their protozoan host. The atomic model reveals both a mostly negatively charged capsid interior, which is conducive to movement of the loosely packed genome, and channels at the 5-fold vertices, which we suggest as routes of mRNA release during transcription. Structural comparison of TVV2 to the Saccharomyces cerevisiae L-A virus reveals a conserved helix-rich fold within the CSP and putative guanylyltransferase domain along the capsid exterior, suggesting conserved mRNA maintenance strategies among Totiviridae This first atomic structure of a TVV provides a framework to guide future biochemical investigations into the interplay between Trichomonas vaginalis and its viruses.IMPORTANCETrichomonas vaginalis viruses (TVVs) are double-stranded RNA (dsRNA) viruses that cohabitate in Trichomonas vaginalis, the causative pathogen of trichomoniasis, the most common nonviral sexually transmitted disease worldwide. Featuring an unsegmented dsRNA genome encoding a single capsid shell protein (CSP), TVVs contrast with multisegmented dsRNA viruses, such as the diarrhea-causing rotavirus, whose larger genome is split into 10 dsRNA segments encoding 5 unique capsid proteins. To determine how TVVs incorporate the requisite functionalities for viral replication into their limited proteome, we derived the atomic model of TVV2, a first for TVVs. Our results reveal the intersubunit interactions driving CSP association for capsid assembly and the properties that govern organization and maintenance of the viral genome. Structural comparison between TVV2 capsids and those of distantly related dsRNA viruses indicates conserved strategies of nascent RNA release and a putative viral guanylyltransferase domain implicated in the cytoplasmic maintenance of viral messenger and genomic RNA.
Keywords: Totiviridae; Trichomonas vaginalis; cryoEM; double-stranded RNA virus; subparticle reconstruction.
Copyright © 2021 Stevens et al.
Figures


Similar articles
-
Trichomonas vaginalis virus: a review of the literature.Int J STD AIDS. 2019 Apr;30(5):496-504. doi: 10.1177/0956462418809767. Epub 2019 Jan 9. Int J STD AIDS. 2019. PMID: 30626281
-
Detection and molecular characterization of double-stranded RNA viruses in Philippine Trichomonas vaginalis isolates.J Microbiol Immunol Infect. 2017 Oct;50(5):669-676. doi: 10.1016/j.jmii.2015.07.016. Epub 2015 Sep 18. J Microbiol Immunol Infect. 2017. PMID: 26471924
-
Genetic characterization of three Cuban Trichomonas vaginalis virus. Phylogeny of Totiviridae family.Infect Genet Evol. 2012 Jan;12(1):113-20. doi: 10.1016/j.meegid.2011.10.020. Epub 2011 Oct 31. Infect Genet Evol. 2012. PMID: 22075038
-
Structure of a protozoan virus from the human genitourinary parasite Trichomonas vaginalis.mBio. 2013 Apr 2;4(2):e00056-13. doi: 10.1128/mBio.00056-13. mBio. 2013. PMID: 23549915 Free PMC article.
-
Trichomonas vaginalis Virus: Current Insights and Emerging Perspectives.Viruses. 2025 Jun 26;17(7):898. doi: 10.3390/v17070898. Viruses. 2025. PMID: 40733518 Free PMC article. Review.
Cited by
-
High-resolution comparative atomic structures of two Giardiavirus prototypes infecting G. duodenalis parasite.PLoS Pathog. 2024 Apr 10;20(4):e1012140. doi: 10.1371/journal.ppat.1012140. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38598600 Free PMC article.
-
Structure of the T=13 capsid of infectious pancreatic necrosis virus (IPNV)-a salmonid birnavirus.J Virol. 2025 Feb 25;99(2):e0145424. doi: 10.1128/jvi.01454-24. Epub 2025 Jan 16. J Virol. 2025. PMID: 39817769 Free PMC article.
-
Microbial Matryoshka: Addressing the Relationship between Pathogenic Flagellated Protozoans and Their RNA Viral Endosymbionts (Family Totiviridae).Vet Sci. 2024 Jul 17;11(7):321. doi: 10.3390/vetsci11070321. Vet Sci. 2024. PMID: 39058005 Free PMC article. Review.
-
Structures of Native Doublet Microtubules from Trichomonas vaginalis Reveal Parasite-Specific Proteins as Potential Drug Targets.Res Sq [Preprint]. 2024 Sep 2:rs.3.rs-4632384. doi: 10.21203/rs.3.rs-4632384/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Apr 29;16(1):3996. doi: 10.1038/s41467-025-59369-y. PMID: 39281863 Free PMC article. Updated. Preprint.
-
Double-Stranded RNA Viruses Are Released From Trichomonas vaginalis Inside Small Extracellular Vesicles and Modulate the Exosomal Cargo.Front Microbiol. 2022 May 4;13:893692. doi: 10.3389/fmicb.2022.893692. eCollection 2022. Front Microbiol. 2022. PMID: 35602021 Free PMC article.
References
-
- World Health Organization. 2012. Global incidence and prevalence of selected curable sexually transmitted infections. World Health Organization.
-
- Stark JR, Judson G, Alderete JF, Mundodi V, Kucknoor AS, Giovannucci EL, Platz EA, Sutcliffe S, Fall K, Kurth T, Ma J, Stampfer MJ, Mucci LA. 2009. Prospective study of Trichomonas vaginalis infection and prostate cancer incidence and mortality: Physicians’ Health Study. J Natl Cancer Inst 101:1406–1411. doi:10.1093/jnci/djp306. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

