Machine learning is the key to diagnose COVID-19: a proof-of-concept study
- PMID: 33785852
- PMCID: PMC8009887
- DOI: 10.1038/s41598-021-86735-9
Machine learning is the key to diagnose COVID-19: a proof-of-concept study
Erratum in
-
Author Correction: Machine learning is the key to diagnose COVID-19: a proof-of-concept study.Sci Rep. 2021 Aug 27;11(1):17577. doi: 10.1038/s41598-021-97049-1. Sci Rep. 2021. PMID: 34453105 Free PMC article. No abstract available.
Abstract
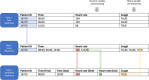
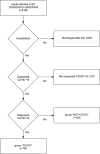
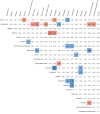
The reverse transcription-polymerase chain reaction (RT-PCR) assay is the accepted standard for coronavirus disease 2019 (COVID-19) diagnosis. As any test, RT-PCR provides false negative results that can be rectified by clinicians by confronting clinical, biological and imaging data. The combination of RT-PCR and chest-CT could improve diagnosis performance, but this would requires considerable resources for its rapid use in all patients with suspected COVID-19. The potential contribution of machine learning in this situation has not been fully evaluated. The objective of this study was to develop and evaluate machine learning models using routine clinical and laboratory data to improve the performance of RT-PCR and chest-CT for COVID-19 diagnosis among post-emergency hospitalized patients. All adults admitted to the ED for suspected COVID-19, and then hospitalized at Rennes academic hospital, France, between March 20, 2020 and May 5, 2020 were included in the study. Three model types were created: logistic regression, random forest, and neural network. Each model was trained to diagnose COVID-19 using different sets of variables. Area under the receiving operator characteristics curve (AUC) was the primary outcome to evaluate model's performances. 536 patients were included in the study: 106 in the COVID group, 430 in the NOT-COVID group. The AUC values of chest-CT and RT-PCR increased from 0.778 to 0.892 and from 0.852 to 0.930, respectively, with the contribution of machine learning. After generalization, machine learning models will allow increasing chest-CT and RT-PCR performances for COVID-19 diagnosis.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

