Schlafen 11 predicts response to platinum-based chemotherapy in gastric cancers
- PMID: 33785877
- PMCID: PMC8257722
- DOI: 10.1038/s41416-021-01364-3
Schlafen 11 predicts response to platinum-based chemotherapy in gastric cancers
Abstract
Background: Although unresectable or recurrent gastric cancers (GC) are frequently treated with platinum-based chemotherapy, response to treatment remains unpredictable. Because Schlafen 11 (SLFN11) is recently identified as a critical determinant of platinum sensitivity, we investigated the potential clinical utility of SLFN11 in the treatment of GC.
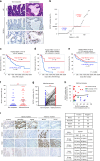
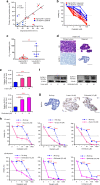
Methods: We analysed the correlation between SLFN11 expression and overall survival in 169 GC patients by our established immunohistochemical approach. The impact of SLFN11 expression on the response to platinum and transition of SLFN11 expression upon long-term treatment with platinum were examined using GC cell lines and organoids.
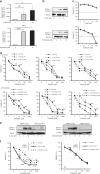
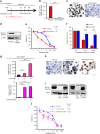
Results: GC patients with high-SLFN11 expression exhibited significantly better survival than those with low-SLFN11 expression, and the significance increased when we selected patients treated with platinum-based chemotherapy. Knockout of SLFN11 and reactivation of SLFN11 in GC cells conferred resistance and sensitivity to platinum, respectively. In GC cells and organoids, long-term treatment with oxaliplatin suppressed SLFN11 expression while imparting drug resistance. The acquired resistance to oxaliplatin was reversed by reactivation of SLFN11 with epigenetic modifying drugs.
Conclusions: This is the first report revealing definitive clinical implications of SLFN11 in the treatment of GC patients and providing novel strategies for the drug selection based on SLFN11 expression.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

