ClassicalGSG: Prediction of log P using classical molecular force fields and geometric scattering for graphs
- PMID: 33786857
- PMCID: PMC8062296
- DOI: 10.1002/jcc.26519
ClassicalGSG: Prediction of log P using classical molecular force fields and geometric scattering for graphs
Abstract
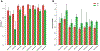
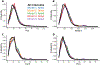
This work examines methods for predicting the partition coefficient (log P) for a dataset of small molecules. Here, we use atomic attributes such as radius and partial charge, which are typically used as force field parameters in classical molecular dynamics simulations. These atomic attributes are transformed into index-invariant molecular features using a recently developed method called geometric scattering for graphs (GSG). We call this approach "ClassicalGSG" and examine its performance under a broad range of conditions and hyperparameters. We train ClassicalGSG log P predictors with neural networks using 10,722 molecules from the OpenChem dataset and apply them to predict the log P values from four independent test sets. The ClassicalGSG method's performance is compared to a baseline model that employs graph convolutional networks. Our results show that the best prediction accuracies are obtained using atomic attributes generated with the CHARMM generalized force field and 2D molecular structures.
Keywords: geometric scattering for graphs; graph convolutional networks; log P prediction; partition coefficients.
© 2021 Wiley Periodicals LLC.
Figures





References
-
- Lipinski CA, Lombardo F, Dominy BW, and Feeney PJ, Advanced Drug Delivery Reviews 23, 3 (1997). - PubMed
-
- Kwon Y, Handbook of essential pharmacokinetics, pharmacodynamics and drug metabolism for industrial scientists (Springer Science & Business Media, 2001).
-
- Ran Y and Yalkowsky SH, Journal of Chemical Information and Computer Sciences 41, 354 (2001). - PubMed
-
- Yalkowsky SH and Valvani SC, Journal of Pharmaceutical Sciences 69, 912 (1980). - PubMed
-
- Ryckmans T, Edwards MP, Horne VA, Correia AM, Owen DR, Thompson LR, Tran I, Tutt MF, and Young T, Bioorganic & Medicinal Chemistry Letters 19, 4406 (2009). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

