Immunity proteins of dual nuclease T6SS effectors function as transcriptional repressors
- PMID: 33786997
- PMCID: PMC8183406
- DOI: 10.15252/embr.202051857
Immunity proteins of dual nuclease T6SS effectors function as transcriptional repressors
Erratum in
-
Immunity proteins of dual nuclease T6SS effectors function as transcriptional repressors.EMBO Rep. 2021 Jun 4;22(6):e53112. doi: 10.15252/embr.202153112. Epub 2021 May 31. EMBO Rep. 2021. PMID: 34060187 Free PMC article.
Abstract
Bacteria utilize type VI secretion system (T6SS) to deliver antibacterial toxins to target co-habiting bacteria. Here, we report that Burkholderia gladioli strain NGJ1 deploys certain T6SS effectors (TseTBg), having both DNase and RNase activities to kill target bacteria. RNase activity is prominent on NGJ1 as well as other bacterial RNA while DNase activity is pertinent to only other bacteria. The associated immunity (TsiTBg) proteins harbor non-canonical helix-turn-helix motifs and demonstrate transcriptional repression activity, similar to the antitoxins of type II toxin-antitoxin (TA) systems. Genome analysis reveals that homologs of TseTBg are either encoded as TA or T6SS effectors in diverse bacteria. Our results indicate that a new ORF (encoding a hypothetical protein) has evolved as a result of operonic fusion of TA type TseTBg homolog with certain T6SS-related genes by the action of IS3 transposable elements. This has potentially led to the conversion of a TA into T6SS effector in Burkholderia. Our study exemplifies that bacteria can recruit toxins of TA systems as T6SS weapons to diversify its arsenal to dominate during inter-bacterial competitions.
Keywords: DNA adenine methylase; LysR proteins; effector neutralization; protein-DNA interaction; restriction modification system.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
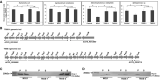
- A
Antibacterial activity of NGJ1, T6SS‐1−, and T6SS‐2− strains against target bacteria.
- B
Schematic representation of two different T6SS apparatus‐encoding gene clusters (named as T6SS‐1 and T6SS‐2) of NGJ1. The locus id (as per Burkholderia genome database) of upstream and downstream genes of the T6SS locus is provided. The gene disrupted in the respective T6SS mutants is marked by triangle (△).
- C, D
Immunoblots showing secretion profile of Hcp‐1 (associated with T6SS‐1) and Hcp‐2 (associated with T6SS‐2) proteins, using protein‐specific peptide antibody in different NGJ1 strains. The T6SS‐1− mutant was defective in Hcp‐1 secretion (due to polar effect of plasmid integration, synthesis was also prevented) but proficient in Hcp‐2 secretion. On the other hand, the T6SS‐2− mutant was defective in secretion of Hcp‐2 (although the protein was synthesized) but proficient in secretion of Hcp‐1.
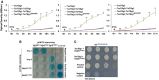
Growth curve of recombinant BL21 (DE3) cells upon IPTG‐mediated induction of proteins, represented as colored line graphs.
Bacterial two‐hybrid assay reflecting interaction between proteins of T6SS apparatus and effector operons. Interaction between T25‐zip and T18‐zip was used as positive control while pKNT25 and pUT18C (empty vectors) were used as negative control. Appearance of blue color suggests positive interaction while the absence of color suggests lack of interaction.
Yeast two‐hybrid assay reflecting interaction of TseTBg1 and TseTBg2 effector proteins with the corresponding PAAR (PAARTseTBg) proteins encoded in their operon. Interaction between P53 and SV40 large T‐antigen (T) proteins was used as positive control. The pGBKT7 and pGADT7 (empty vectors) were used as negative control.

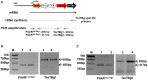
Strategy used for validating the expression of different genes of TseTBg operon as single transcriptional unit.
PCR amplification of PAARTseTBg1 and TseTBg1 genes from cDNA synthesized using TsiTBg1‐specific primer.
PCR amplification of PAARTseTBg2 and TseTBg2 genes from cDNA synthesized using TsiTBg2‐specific primer.
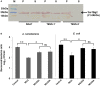
Western blot analysis reflecting the T6SS‐2‐dependent secretion of TseTBg2 protein in NGJ1. The total proteins from cell‐free supernatant (S) as well as pellet (P) of different strains were immunoblotted using TseTBg2‐specific peptide antibody.
Antibacterial activity of the TseTBg2 mutants (NGJ203 and NGJ204; two independent mutants) and wild‐type NGJ1 bacteria against Agrobacterium tumefaciens and Escherichia coli.
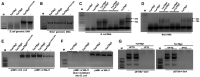
- A, B
Effect of purified TseTBg (TseTBg1/TseTBg2) and one of the variants (TseTBg2D116A,K129A) proteins on degradation of bacterial genomic DNA.
- C, D
RNase activity of TseTBg effector proteins, in the presence or absence of cognate TsiTBg immunity proteins.
- E
Effect of TseTBg1, TseTBg2, TseTBg2D116A,K129A proteins on degradation of pHM1, a broad host‐range plasmid isolated from Escherichia coli and NGJ1.
- F
Effect of TseTBg1 and TseTBg2 proteins on degradation of pHM1, reintroduced into E. coli from NGJ1.
- G
Effect of ectopic expression of DNA adenine methylase (Dam) protein on TseTBg proteins mediated degradation of plasmid DNA.

Growth of recombinant BL21 (DE3) cells with or without induction of TseTBg2 and its variant (TseTBg2D116A,K129A, DNase−RNase+).
Growth curve of recombinant BL21 (DE3) cells with or without IPTG‐mediated induction of TseTBg2 or TseTBg2D116A,K129A proteins.

- A, B
Schematics reflecting the presence of a non‐canonical HTH domain and an Imm52 domain in TsiTBg1 and TsiTBg2 proteins.
- C, D
Representation of predicted external (pExtTseTBg1/pExtTseTBg2) and internal (pIntTseTBg1/pIntTseTBg2) promoter regions in TseTBg1 and TseTBg2 operons. Strategy for obtaining promoter:Gus reporter constructs in pBI101 is also provided.
- E, F
Graph showing the effect of native TsiTBg and HTH or Imm52 domain‐deleted variant of TsiTBg proteins on Gus reporter gene expression (quantified using 4‐methylumbelliferyl ß‐
d ‐glucuronide, MUG, as substrate) under the external (pExtTseTBg1/pExtTseTBg2) and internal (pIntTseTBg1/pIntTseTBg2) promoters in Escherichia coli.
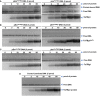
Interaction of external (pExtTseTBg1) and internal (pIntTseTBg1) promoter DNA fragments of TseTBg1 operon with the TsiTBg1 protein.
Interaction of pExtTseTBg1 and pIntTseTBg1 promoter DNA fragments of TseTBg1 operon with the HTH domain‐deleted variant of TseTBg1 protein (TsiTBg1ΔHTH).
Interaction of pExtTseTBg2 and pIntTseTBg2 promoter DNA of TseTBg2 operon with the TsiTBg1 protein.
Interaction of control (random) DNA with the TsiTBg1 protein.

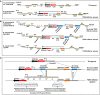
The genomic loci containing TseTBg (toxin) and TsiTBg (antitoxin) homologs in different B. pseudomallei strains.
In B. pseudomallei 3000015237 isolate MX2014, two IS3 family transposable elements that carry two of the T6SS‐related genes (PAAR and DUF4123) are present upstream to the TA genes. However, in B. pseudomallei Nau24B‐3 and vgh16R strains, the IS3 element which flank the TA and T6SS‐related genes have been excised and a new ORF encoding hypothetical protein (H.P) has evolved at its place.
The excision of IS3 family transposase has created operonic fusion of TA and T6SS related genes. Moreover, a new ORF of 474 bp that encode a H.P has evolved at the locus, through adoption of 3′ end of the DUF4123 and 5′ end of the toxin gene.

References
-
- Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ (2017) Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21: 769–776.e3 - PubMed
-
- Benz J, Meinhart A (2014) Antibacterial effector/immunity systems: it's just the tip of the iceberg. Curr Opin Microbiol 17: 1–10 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

