Functional characterization of PD1+TIM3+ tumor-infiltrating T cells in DLBCL and effects of PD1 or TIM3 blockade
- PMID: 33787861
- PMCID: PMC8045516
- DOI: 10.1182/bloodadvances.2020003080
Functional characterization of PD1+TIM3+ tumor-infiltrating T cells in DLBCL and effects of PD1 or TIM3 blockade
Abstract
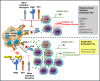
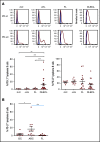
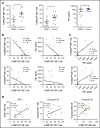
In diffuse large B-cell lymphoma (DLBCL), tumor-infiltrating T lymphocytes (TILs) are involved in therapeutic responses. However, tumor-specific TILs can be dysfunctional, with impaired effector functions. Various mechanisms are involved in this exhaustion, and the increased expression of programmed cell death receptor 1 (PD1) and TIM3 on dysfunctional cells suggests their involvement. However, conflicting data have been published regarding their expression or coexpression in DLBCL. We evaluated the presence and phenotype of CD4+ and CD8+ TILs in freshly collected tumor tissues in DLBCL and compared the results with those in follicular lymphoma, classical Hodgkin lymphoma, and nonmalignant reactive lymphadenopathy. We found that TILs expressing both PD1 and TIM3 were expanded in DLBCL, particularly in the activated B cell-like subgroup. Isolated PD1+TIM3+ TILs exhibited a transcriptomic signature related to T-cell exhaustion associated with a reduction in cytokine production, both compromising the antitumor immune response. However, these cells expressed high levels of cytotoxic molecules. In line with this, stimulated PD1+TIM3+ TILs from DLBCL patients exhibited reduced proliferation and impaired secretion of interferon-γ, but these functions were restored by the blockade of PD1 or TIM3. In summary, the PD1+TIM3+ TIL population is expanded and exhausted in DLBCL but can be reinvigorated with appropriate therapies.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: D.O. is cofounder of ImCheck Therapeutics, Alderaan Biotechnology, and Emergence Therapeutics and is a shareholder in these companies. The remaining authors declare no competing financial interests.
Figures






References
-
- Staiger AM, Ziepert M, Horn H, et al. ; German High-Grade Lymphoma Study Group . Clinical impact of the cell-of-origin classification and the MYC/ BCL2 dual expresser status in diffuse large B-cell lymphoma treated within prospective clinical trials of the German High-Grade Non-Hodgkin’s Lymphoma Study Group. J Clin Oncol. 2017;35(22):2515-2526. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

