Bacteria primed by antimicrobial peptides develop tolerance and persist
- PMID: 33788905
- PMCID: PMC8041211
- DOI: 10.1371/journal.ppat.1009443
Bacteria primed by antimicrobial peptides develop tolerance and persist
Abstract
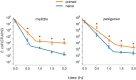
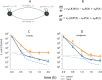
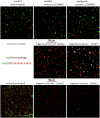
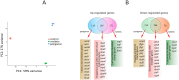
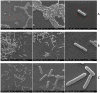
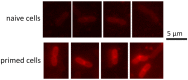
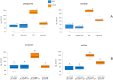
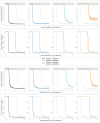
Antimicrobial peptides (AMPs) are key components of innate immune defenses. Because of the antibiotic crisis, AMPs have also come into focus as new drugs. Here, we explore whether prior exposure to sub-lethal doses of AMPs increases bacterial survival and abets the evolution of resistance. We show that Escherichia coli primed by sub-lethal doses of AMPs develop tolerance and increase persistence by producing curli or colanic acid, responses linked to biofilm formation. We develop a population dynamic model that predicts that priming delays the clearance of infections and fuels the evolution of resistance. The effects we describe should apply to many AMPs and other drugs that target the cell surface. The optimal strategy to tackle tolerant or persistent cells requires high concentrations of AMPs and fast and long-lasting expression. Our findings also offer a new understanding of non-inherited drug resistance as an adaptive response and could lead to measures that slow the evolution of resistance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Koo HB, Seo J. Antimicrobial peptides under clinical investigation. Pept Sci. John Wiley and Sons Inc.; 2019;111. 10.1002/pep2.24122 - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

