SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera
- PMID: 33789085
- PMCID: PMC7980135
- DOI: 10.1016/j.chom.2021.03.008
SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera
Abstract
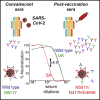
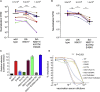
Toward eradicating the COVID-19 pandemic, vaccines that induce high humoral and cellular immune responses are essential. However, SARS-CoV-2 variants have begun to emerge and raise concerns, as they may potentially compromise vaccine efficiency. Here, we monitored neutralization potency of convalescent or Pfizer-BTN162b2 post-vaccination sera against pseudoviruses displaying spike proteins derived from wild-type SARS-CoV-2, or its UK-B.1.1.7 and SA-B.1.351 variants. Compared to convalescent sera, vaccination induces high titers of neutralizing antibodies, which exhibit efficient neutralization potential against pseudovirus carrying wild-type SARS-CoV-2. However, while wild-type and UK-N501Y pseudoviruses were similarly neutralized, those displaying SA-N501Y/K417N/E484K spike mutations moderately resist neutralization. Contribution of single or combined spike mutations to neutralization and infectivity were monitored, highlighting mechanisms by which viral infectivity and neutralization resistance are enhanced by N501Y or E484K/K417N mutations. Our study validates the importance of the Pfizer vaccine but raises concerns regarding its efficacy against specific SARS-CoV-2 circulating variants.
Keywords: COVID-19; Pfizer-BTN162b2 vaccine; SARS-CoV-2; Spike; UK and SA variants; United Kingdom and South African variants; neutralization antibodies.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Annavajhala M.K., Mohri H., Zucker J.E., Sheng Z., Wang P., Gomez-Simmonds A., Ho D.D., Uhlemann A.-C. A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv. 2021 doi: 10.1101/2021.02.23.21252259. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

