Alterations in the human oral and gut microbiomes and lipidomics in COVID-19
- PMID: 33789966
- PMCID: PMC8042598
- DOI: 10.1136/gutjnl-2020-323826
Alterations in the human oral and gut microbiomes and lipidomics in COVID-19
Abstract
Objective: To characterise the oral microbiome, gut microbiome and serum lipid profiles in patients with active COVID-19 and recovered patients; evaluate the potential of the microbiome as a non-invasive biomarker for COVID-19; and explore correlations between the microbiome and lipid profile.
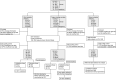
Design: We collected and sequenced 392 tongue-coating samples, 172 faecal samples and 155 serum samples from Central China and East China. We characterised microbiome and lipid molecules, constructed microbial classifiers in discovery cohort and verified their diagnostic potential in 74 confirmed patients (CPs) from East China and 37 suspected patients (SPs) with IgG positivity.
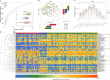
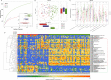
Results: Oral and faecal microbial diversity was significantly decreased in CPs versus healthy controls (HCs). Compared with HCs, butyric acid-producing bacteria were decreased and lipopolysaccharide-producing bacteria were increased in CPs in oral cavity. The classifiers based on 8 optimal oral microbial markers (7 faecal microbial markers) achieved good diagnostic efficiency in different cohorts. Importantly, diagnostic efficacy reached 87.24% in the cross-regional cohort. Moreover, the classifiers successfully diagnosed SPs with IgG antibody positivity as CPs, and diagnostic efficacy reached 92.11% (98.01% of faecal microbiome). Compared with CPs, 47 lipid molecules, including sphingomyelin (SM)(d40:4), SM(d38:5) and monoglyceride(33:5), were depleted, and 122 lipid molecules, including phosphatidylcholine(36:4p), phosphatidylethanolamine (PE)(16:0p/20:5) and diglyceride(20:1/18:2), were enriched in confirmed patients recovery.
Conclusion: This study is the first to characterise the oral microbiome in COVID-19, and oral microbiomes and lipid alterations in recovered patients, to explore their correlations and to report the successful establishment and validation of a diagnostic model for COVID-19.
Keywords: COVID-19; intestinal microbiology; lipid metabolism.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures





References
-
- WHO . Coronavirus disease (COVID-19) Weekly epidemiological update and Weekly operational update, 2020. Available: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio...
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
