Pan-ERBB kinase inhibition augments CDK4/6 inhibitor efficacy in oesophageal squamous cell carcinoma
- PMID: 33789967
- PMCID: PMC8921580
- DOI: 10.1136/gutjnl-2020-323276
Pan-ERBB kinase inhibition augments CDK4/6 inhibitor efficacy in oesophageal squamous cell carcinoma
Erratum in
-
Correction: Pan-ERBB kinase inhibition augments CDK4/6 inhibitor efficacy in oesophageal squamous cell carcinoma.Gut. 2023 Nov;72(11):e5. doi: 10.1136/gutjnl-2020-323276corr1. Gut. 2023. PMID: 37802538 Free PMC article. No abstract available.
Abstract
Objective: Oesophageal squamous cell carcinoma (OSCC), like other squamous carcinomas, harbour highly recurrent cell cycle pathway alterations, especially hyperactivation of the CCND1/CDK4/6 axis, raising the potential for use of existing CDK4/6 inhibitors in these cancers. Although CDK4/6 inhibition has shown striking success when combined with endocrine therapy in oestrogen receptor positive breast cancer, CDK4/6 inhibitor palbociclib monotherapy has not revealed evidence of efficacy to date in OSCC clinical studies. Herein, we sought to elucidate the identification of key dependencies in OSCC as a foundation for the selection of targets whose blockade could be combined with CDK4/6 inhibition.
Design: We combined large-scale genomic dependency and pharmaceutical screening datasets with preclinical cell line models, to identified potential combination therapies in squamous cell cancer.
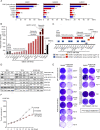
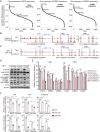
Results: We identified sensitivity to inhibitors to the ERBB family of receptor kinases, results clearly extending beyond the previously described minority of tumours with EGFR amplification/dependence, specifically finding a subset of OSCCs with dual dependence on ERBB3 and ERBB2. Subsequently. we demonstrated marked efficacy of combined pan-ERBB and CDK4/6 inhibition in vitro and in vivo. Furthermore, we demonstrated that squamous lineage transcription factor KLF5 facilitated activation of ERBBs in OSCC.
Conclusion: These results provide clear rationale for development of combined ERBB and CDK4/6 inhibition in these cancers and raises the potential for KLF5 expression as a candidate biomarker to guide the use of these agents. These data suggested that by combining existing Food and Drug Administration (FDA)-approved agents, we have the capacity to improve therapy for OSCC and other squamous cancer.
Keywords: cell cycle control; oesophageal cancer; pharmacogenomics; pharmacotherapy.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: AB receives research funding from Bayer, Merck and Novartis and is a consultant to Earli, and HelixNano and a cofounder of Signet Therapeutics. K-KW is a founder and equity holder of G1 Therapeutics, and he has consulting/sponsored research agreements with MedImmune, Takeda, TargImmune, BMS, AstraZeneca, Janssen, Pfizer, Novartis, Merck, Ono and Array.
Figures


References
-
- Kato K, Cho BC, Takahashi M, et al. Nivolumab versus chemotherapy in patients with advanced oesophageal squamous cell carcinoma refractory or intolerant to previous chemotherapy (ATTRACTION-3): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol 2019;20:1506–17. 10.1016/S1470-2045(19)30626-6 - DOI - PubMed
-
- Shah MA, Kojima T, Hochhauser D, et al. Efficacy and safety of pembrolizumab for heavily pretreated patients with advanced, metastatic adenocarcinoma or squamous cell carcinoma of the esophagus: the phase 2 KEYNOTE-180 study. JAMA Oncol 2019;5:546–50. 10.1001/jamaoncol.2018.5441 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
