Validation of an Isothermal Amplification Platform for Microbial Identification and Antimicrobial Resistance Detection in Blood: A Prospective Study
- PMID: 33790511
- PMCID: PMC7991769
- DOI: 10.5005/jp-journals-10071-23761
Validation of an Isothermal Amplification Platform for Microbial Identification and Antimicrobial Resistance Detection in Blood: A Prospective Study
Abstract
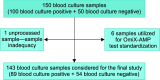
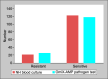
Background: Recent advances in nucleic acid amplification technique (NAAT)-based identification of pathogens in blood stream infections (BSI) have revolutionized molecular diagnostics in comparison to traditional clinical microbiology practice of blood culture. Rapid pathogen detection with point-of-care diagnostic-applicable platform is prerequisite for efficient patient management. The aim of the study is to evaluate an in-house developed, lyophilized OmiX-AMP pathogen test for the detection of top six BSI-causing bacteria along with two major antimicrobial resistance (AMR) markers of carbapenem and compare it to the traditional blood culture-based detection. Materials and methods: One hundred forty-three patients admitted to the Medical Intensive Care Unit, Narayana Hrudayalaya, Bangalore, with either suspected or proven sepsis, of either gender, of age ≥18 years were enrolled for the study. Pathogen DNA extracted from blood culture sample using OmiX pReP method was amplified at isothermal conditions and analyzed in real time using OmiX Analysis software. Results: Among the processed 143 samples, 54 were true negative, 83 were true positive, 3 were false negative, and 2 were false positive as analyzed by OmiX READ software. Gram-negative bacteria (91.3%) and gram-positive bacteria (75%) were detected with 100% specificity and 95.6% sensitivity along with the AMR marker pattern with a turnaround time of 4 hours from sample collection to results. Conclusion: OmiX-AMP pathogen test detected pathogens with 96.5% concordance in comparison to traditional blood culture. Henceforth, OmiX-AMP pathogen test could be used as a readily deployable diagnostic kit even in low-resource settings. How to cite this article: Maheshwarappa HM, Guru P, Mundre RS, Lawrence N, Majumder S, Sigamani A, et al. Validation of an Isothermal Amplification Platform for Microbial Identification and Antimicrobial Resistance Detection in Blood: A Prospective Study. Indian J Crit Care Med 2021;25(3):299-304.
Keywords: Blood stream infection; Diagnosis; Isothermal amplification; Pathogen detection; Sepsis.
Copyright © 2021; Jaypee Brothers Medical Publishers (P) Ltd.
Conflict of interest statement
Source of support: This study was supported by funding provided from from OmiX Research and Diagnostics Laboratories to Narayaha Hrudalaya. The funding sources were grants from BIRAC, Bill and Melinda Gates Foundation, Government of Karnataka Idea2PoC to OmiX Research and Diagnostics Laboratories. Conflict of interest: None
Figures

References
-
- Abubakar II, Tillmann T, Banerjee A. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study. Lancet. 2015;;385::117––171.. doi: 10.1016/S0140-6736(14)61682-2. DOI: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
