Functional Analysis of Wheat NAC Transcription Factor, TaNAC069, in Regulating Resistance of Wheat to Leaf Rust Fungus
- PMID: 33790919
- PMCID: PMC8005738
- DOI: 10.3389/fpls.2021.604797
Functional Analysis of Wheat NAC Transcription Factor, TaNAC069, in Regulating Resistance of Wheat to Leaf Rust Fungus
Abstract
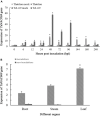
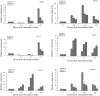
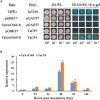
NAC transcription factors are one of the largest transcription factor families having functions in a variety of stress responses. Few NACs have been reported for interactions between wheat and the wheat rust fungus Puccinia triticina (Pt). In this study, based on analysis of RNA-seq data from wheat line TcLr19 inoculated by Pt, the NAC transcription factor TaNAC069 was cloned from wheat, and its transcriptional activity and homologous dimer formation were verified. Quantitative real-time PCR analysis showed that the expression of TaNAC069 was induced by Pt and associated signaling molecules. To further characterize the function of the TaNAC069 gene in wheat resistance to Pt, virus-induced gene silencing (VIGS) was utilized, and it revealed that Pt resistance in TaNAC069-silenced plants was significantly reduced. Potential interaction targets of TaNAC069 from wheat and Pt were screened and identified by yeast two-hybrid technology. Eukaryotic elongation factor eEF1A, CBSX3 protein, and cold acclimation protein WCOR410c were screened by yeast one-hybrid technology. The results indicate that the TaNAC069 gene plays a positive regulatory role in wheat resistance to Pt, laying a good foundation to analyze the molecular mechanisms of TaNAC069 and its functional role in wheat resistance to Pt.
Keywords: NAC transcription factor; Puccinia triticina; RNA-seq; VIGS; clone.
Copyright © 2021 Zhang, Geng, Cui, Wang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

