Automated Coding of Under-Studied Medical Concept Domains: Linking Physical Activity Reports to the International Classification of Functioning, Disability, and Health
- PMID: 33791684
- PMCID: PMC8009547
- DOI: 10.3389/fdgth.2021.620828
Automated Coding of Under-Studied Medical Concept Domains: Linking Physical Activity Reports to the International Classification of Functioning, Disability, and Health
Abstract
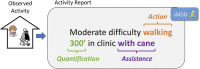
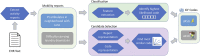
Linking clinical narratives to standardized vocabularies and coding systems is a key component of unlocking the information in medical text for analysis. However, many domains of medical concepts, such as functional outcomes and social determinants of health, lack well-developed terminologies that can support effective coding of medical text. We present a framework for developing natural language processing (NLP) technologies for automated coding of medical information in under-studied domains, and demonstrate its applicability through a case study on physical mobility function. Mobility function is a component of many health measures, from post-acute care and surgical outcomes to chronic frailty and disability, and is represented as one domain of human activity in the International Classification of Functioning, Disability, and Health (ICF). However, mobility and other types of functional activity remain under-studied in the medical informatics literature, and neither the ICF nor commonly-used medical terminologies capture functional status terminology in practice. We investigated two data-driven paradigms, classification and candidate selection, to link narrative observations of mobility status to standardized ICF codes, using a dataset of clinical narratives from physical therapy encounters. Recent advances in language modeling and word embedding were used as features for established machine learning models and a novel deep learning approach, achieving a macro-averaged F-1 score of 84% on linking mobility activity reports to ICF codes. Both classification and candidate selection approaches present distinct strengths for automated coding in under-studied domains, and we highlight that the combination of (i) a small annotated data set; (ii) expert definitions of codes of interest; and (iii) a representative text corpus is sufficient to produce high-performing automated coding systems. This research has implications for continued development of language technologies to analyze functional status information, and the ongoing growth of NLP tools for a variety of specialized applications in clinical care and research.
Keywords: Disability; EHR data; Free text; ICF; Machine learning; Natural language processing; Physical function; Rehabilitation.
Conflict of interest statement
Conflict of Interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Hatef E, Rouhizadeh M, Tia I, Lasser E, Hill-Briggs F, Marsteller J, et al. Assessing the availability of data on social and behavioral determinants in structured and unstructured electronic health records: a retrospective analysis of a multilevel health care system. JMIR Med Inf. (2019) 7:e13802. 10.2196/13802 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

