This is a preprint.
Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution
- PMID: 33791709
- PMCID: PMC8010738
- DOI: 10.1101/2021.03.19.21253391
Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution
Update in
-
Independent infections of porcine deltacoronavirus among Haitian children.Nature. 2021 Dec;600(7887):133-137. doi: 10.1038/s41586-021-04111-z. Epub 2021 Nov 17. Nature. 2021. PMID: 34789872 Free PMC article.
Abstract
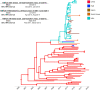
Coronaviruses have caused three major epidemics since 2003, including the ongoing SARS-CoV-2 pandemic. In each case, coronavirus emergence in our species has been associated with zoonotic transmissions from animal reservoirs 1,2 , underscoring how prone such pathogens are to spill over and adapt to new species. Among the four recognized genera of the family Coronaviridae - Alphacoronavirus, Betacoronavirus, Deltacoronavirus, Gammacoronavirus , - human infections reported to date have been limited to alpha- and betacoronaviruses 3 . We identify, for the first time, porcine deltacoronavirus (PDCoV) strains in plasma samples of three Haitian children with acute undifferentiated febrile illness. Genomic and evolutionary analyses reveal that human infections were the result of at least two independent zoonoses of distinct viral lineages that acquired the same mutational signature in the nsp15 and the spike glycoprotein genes by convergent evolution. In particular, structural analysis predicts that one of the changes in the Spike S1 subunit, which contains the receptor-binding domain, may affect protein's flexibility and binding to the host cell receptor. Our findings not only underscore the ability of deltacoronaviruses to adapt and potentially lead to human-to-human transmission, but also raise questions about the role of such transmissions in development of pre-existing immunity to other coronaviruses, such as SARS-CoV-2.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
