This is a preprint.
Femtomolar SARS-CoV-2 Antigen Detection Using the Microbubbling Digital Assay with Smartphone Readout Enables Antigen Burden Quantitation and Dynamics Tracking
- PMID: 33791710
- PMCID: PMC8010739
- DOI: 10.1101/2021.03.17.21253847
Femtomolar SARS-CoV-2 Antigen Detection Using the Microbubbling Digital Assay with Smartphone Readout Enables Antigen Burden Quantitation and Dynamics Tracking
Update in
-
Femtomolar SARS-CoV-2 Antigen Detection Using the Microbubbling Digital Assay with Smartphone Readout Enables Antigen Burden Quantitation and Tracking.Clin Chem. 2021 Dec 30;68(1):230-239. doi: 10.1093/clinchem/hvab158. Clin Chem. 2021. PMID: 34383886 Free PMC article.
Abstract
Background: Little is known about the dynamics of SARS-CoV-2 antigen burden in respiratory samples in different patient populations at different stages of infection. Current rapid antigen tests cannot quantitate and track antigen dynamics with high sensitivity and specificity in respiratory samples.
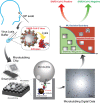
Methods: We developed and validated an ultra-sensitive SARS-CoV-2 antigen assay with smartphone readout using the Microbubbling Digital Assay previously developed by our group, which is a platform that enables highly sensitive detection and quantitation of protein biomarkers. A computer vision-based algorithm was developed for microbubble smartphone image recognition and quantitation. A machine learning-based classifier was developed to classify the smartphone images based on detected microbubbles. Using this assay, we tracked antigen dynamics in serial swab samples from COVID patients hospitalized in ICU and immunocompromised COVID patients.
Results: The limit of detection (LOD) of the Microbubbling SARS-CoV-2 Antigen Assay was 0.5 pg/mL (10.6 fM) recombinant nucleocapsid (N) antigen or 4000 copies/mL inactivated SARS-CoV-2 virus in nasopharyngeal (NP) swabs, comparable to many rRT-PCR methods. The assay had high analytical specificity towards SARS-CoV-2. Compared to EUA-approved rRT-PCR methods, the Microbubbling Antigen Assay demonstrated a positive percent agreement (PPA) of 97% (95% confidence interval (CI), 92-99%) in symptomatic individuals within 7 days of symptom onset and positive SARS-CoV-2 nucleic acid results, and a negative percent agreement (NPA) of 97% (95% CI, 94-100%) in symptomatic and asymptomatic individuals with negative nucleic acid results. Antigen positivity rate in NP swabs gradually decreased as days-after-symptom-onset increased, despite persistent nucleic acid positivity of the same samples. The computer vision and machine learning-based automatic microbubble image classifier could accurately identify positives and negatives, based on microbubble counts and sizes. Total microbubble volume, a potential marker of antigen burden, correlated inversely with Ct values and days-after-symptom-onset. Antigen was detected for longer periods of time in immunocompromised patients with hematologic malignancies, compared to immunocompetent individuals. Simultaneous detectable antigens and nucleic acids may indicate the presence of replicating viruses in patients with persistent infections.
Conclusions: The Microbubbling SARS-CoV-2 Antigen Assay enables sensitive and specific detection of acute infections, and quantitation and tracking of antigen dynamics in different patient populations at various stages of infection. With smartphone compatibility and automated image processing, the assay is well-positioned to be adapted for point-of-care diagnosis and to explore the clinical implications of antigen dynamics in future studies.
Figures



References
-
- SARS-CoV-2 Reference Panel Comparative Data. https://www.fda.gov/medical-https://www.fda.gov/medical-devices/coronavi...
-
- Coronavirus (COVID-19) Update: FDA Authorizes First Antigen Test to Help in the Rapid Detection of the Virus that Causes COVID-19 in Patients., <https://www.fda.gov/news-https://www.fda.gov/news-events/press-announcem...> (2020).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
