Building blocks and blueprints for bacterial autolysins
- PMID: 33793553
- PMCID: PMC8051824
- DOI: 10.1371/journal.pcbi.1008889
Building blocks and blueprints for bacterial autolysins
Abstract
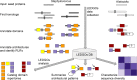
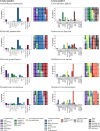
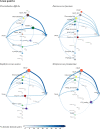
Bacteria utilize a wide variety of endogenous cell wall hydrolases, or autolysins, to remodel their cell walls during processes including cell division, biofilm formation, and programmed death. We here systematically investigate the composition of these enzymes in order to gain insights into their associated biological processes, potential ways to disrupt them via chemotherapeutics, and strategies by which they might be leveraged as recombinant antibacterial biotherapies. To do so, we developed LEDGOs (lytic enzyme domains grouped by organism), a pipeline to create and analyze databases of autolytic enzyme sequences, constituent domain annotations, and architectural patterns of multi-domain enzymes that integrate peptidoglycan binding and degrading functions. We applied LEDGOs to eight pathogenic bacteria, gram negatives Acinetobacter baumannii, Klebsiella pneumoniae, Neisseria gonorrhoeae, and Pseudomonas aeruginosa; and gram positives Clostridioides difficile, Enterococcus faecium, Staphylococcus aureus, and Streptococcus pneumoniae. Our analysis of the autolytic enzyme repertoires of these pathogens reveals commonalities and differences in their key domain building blocks and architectures, including correlations and preferred orders among domains in multi-domain enzymes, repetitions of homologous binding domains with potentially complementarity recognition modalities, and sequence similarity patterns indicative of potential divergence of functional specificity among related domains. We have further identified a variety of unannotated sequence regions within the lytic enzymes that may themselves contain new domains with important functions.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: KEG and CB-K are member-managers of Lyticon LLC. No other authors have a conflict of interest. Potential conflicts of interest for KEG and CB-K are under management at Dartmouth. The authors declare that the work presented here is free of any bias.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

