Using Hawkes Processes to model imported and local malaria cases in near-elimination settings
- PMID: 33793564
- PMCID: PMC8043404
- DOI: 10.1371/journal.pcbi.1008830
Using Hawkes Processes to model imported and local malaria cases in near-elimination settings
Abstract
Developing new methods for modelling infectious diseases outbreaks is important for monitoring transmission and developing policy. In this paper we propose using semi-mechanistic Hawkes Processes for modelling malaria transmission in near-elimination settings. Hawkes Processes are well founded mathematical methods that enable us to combine the benefits of both statistical and mechanistic models to recreate and forecast disease transmission beyond just malaria outbreak scenarios. These methods have been successfully used in numerous applications such as social media and earthquake modelling, but are not yet widespread in epidemiology. By using domain-specific knowledge, we can both recreate transmission curves for malaria in China and Eswatini and disentangle the proportion of cases which are imported from those that are community based.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
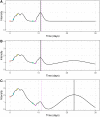
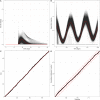
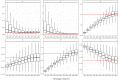

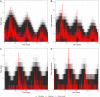

References
-
- Kermack WO, McKendrick AG, Walker GT. A contribution to the mathematical theory of epidemics. Proceedings of the Royal Society of London Series A, Containing Papers of a Mathematical and Physical Character. 1927;115(772):700–721.
-
- Routledge I, Lai S, Battle KE, Ghani AC, Gomez-Rodriguez M, Gustafson KB, et al.. Tracking progress towards malaria elimination in China: Individual-level estimates of transmission and its spatiotemporal variation using a diffusion network approach. PLOS Computational Biology. 2020;16(3):1–20. 10.1371/journal.pcbi.1007707 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

